Short Course on R Tools
Mehdi Maadooliat and Hossein Haghbin
Marquette University
SCoRT - Summer 2025
Motivating Example - What is Data Science?
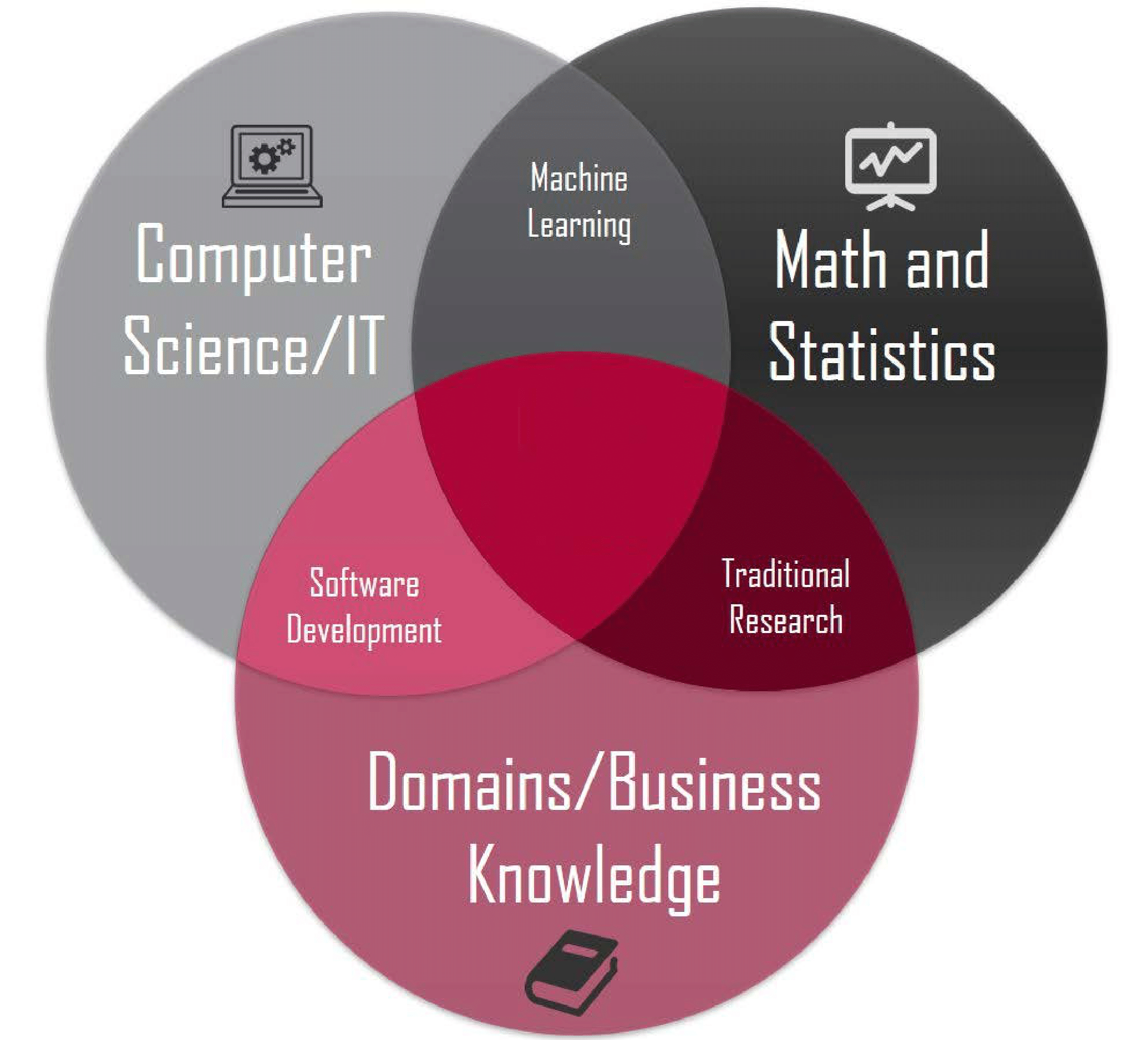
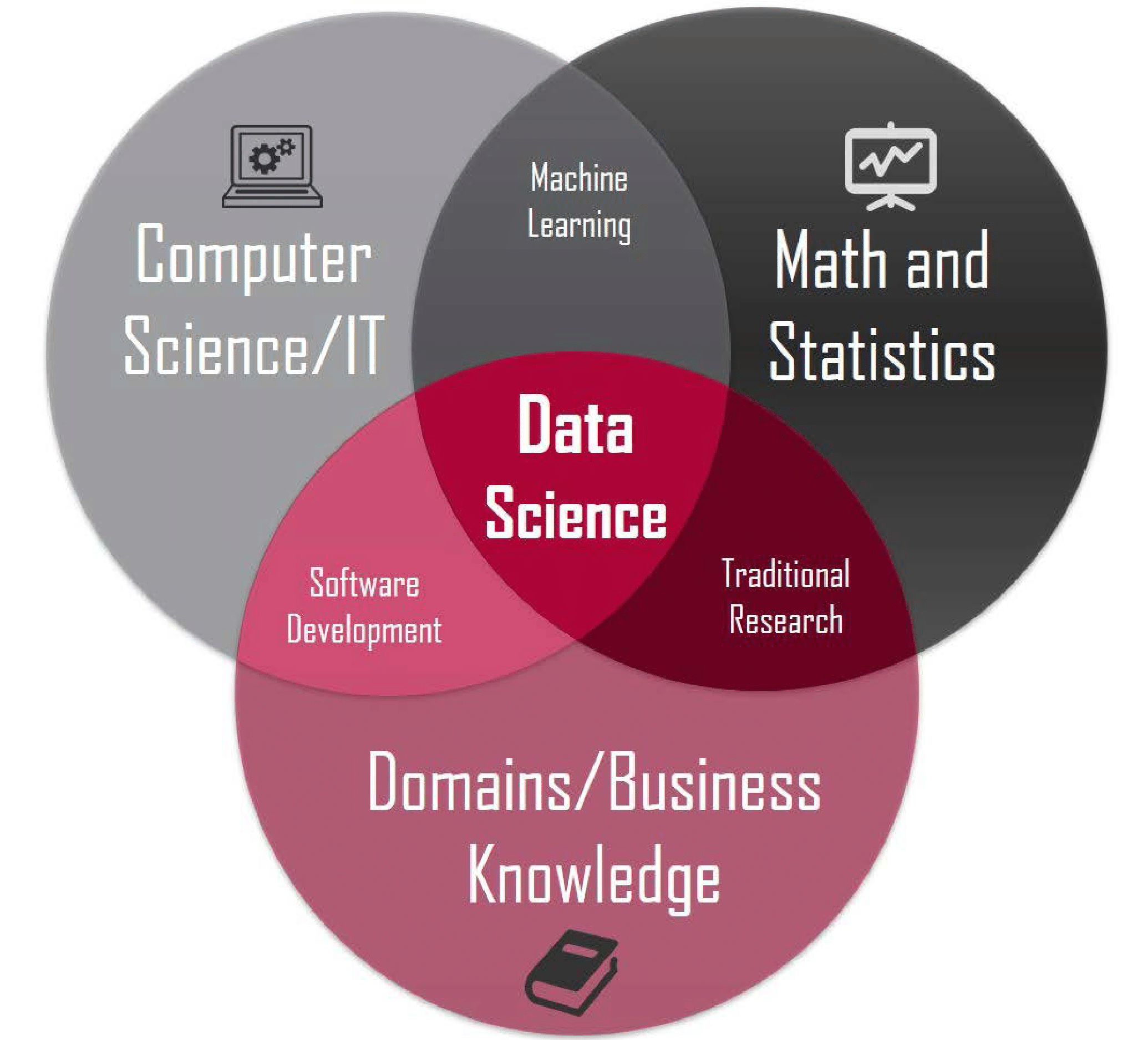
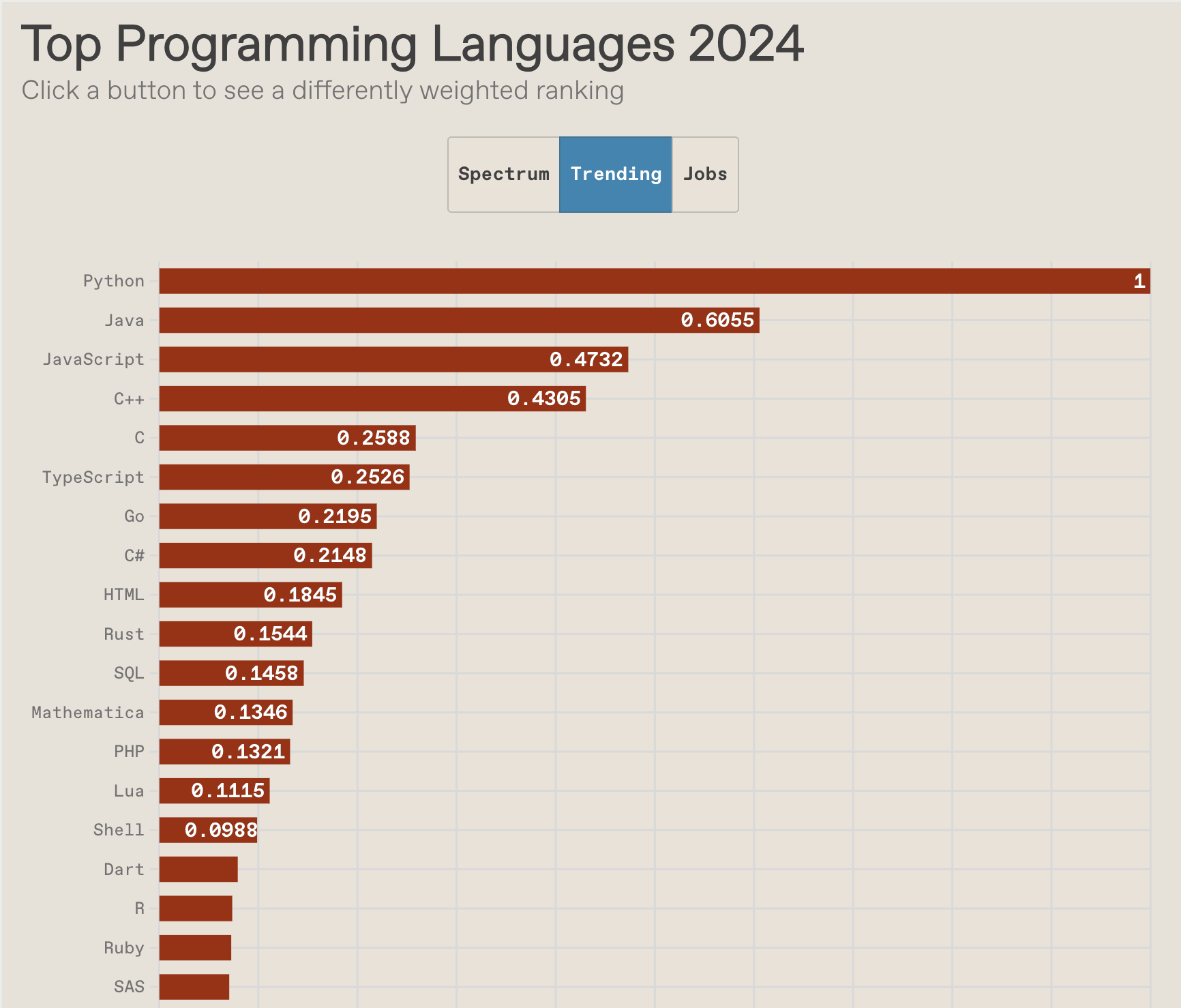
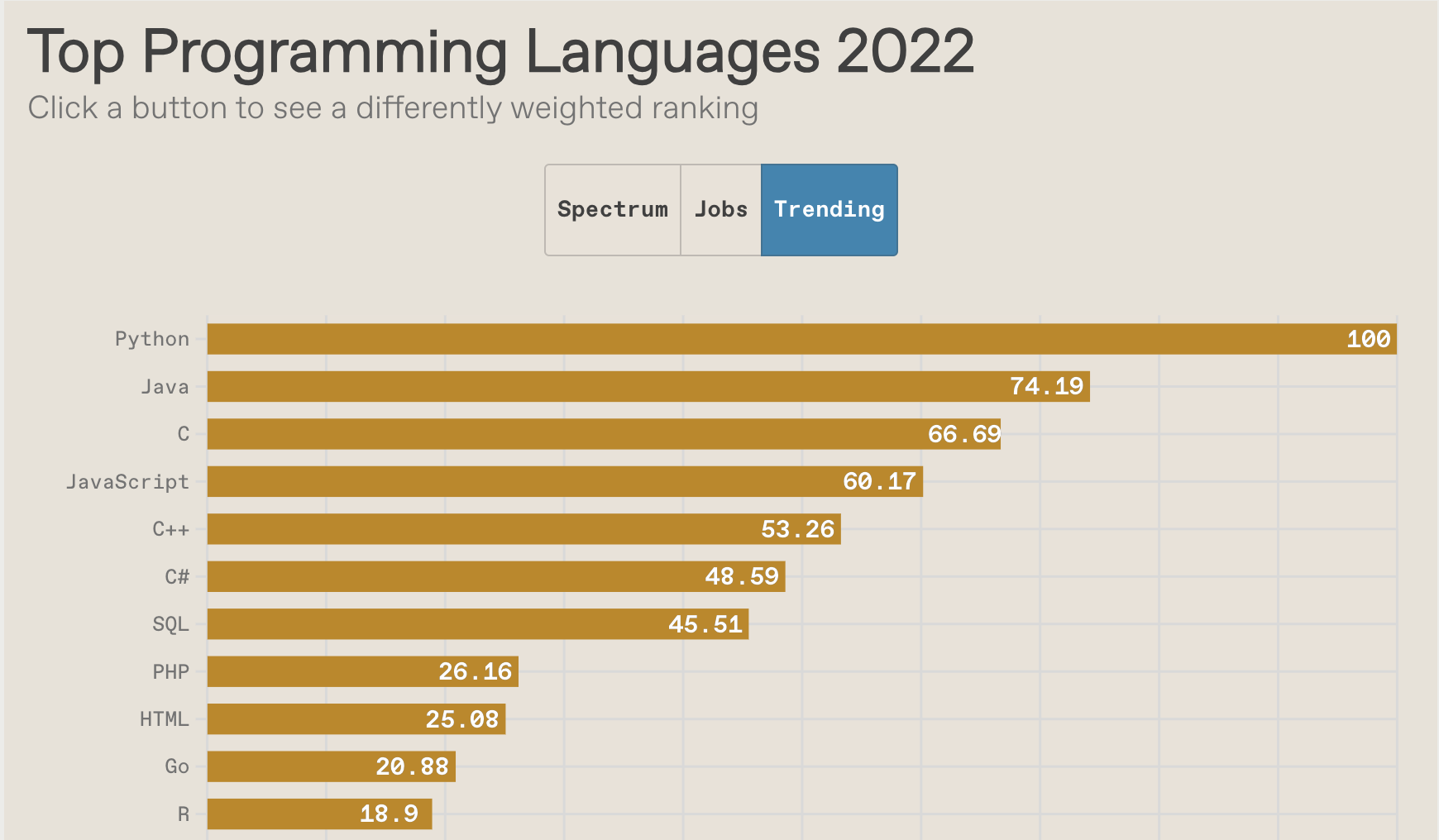
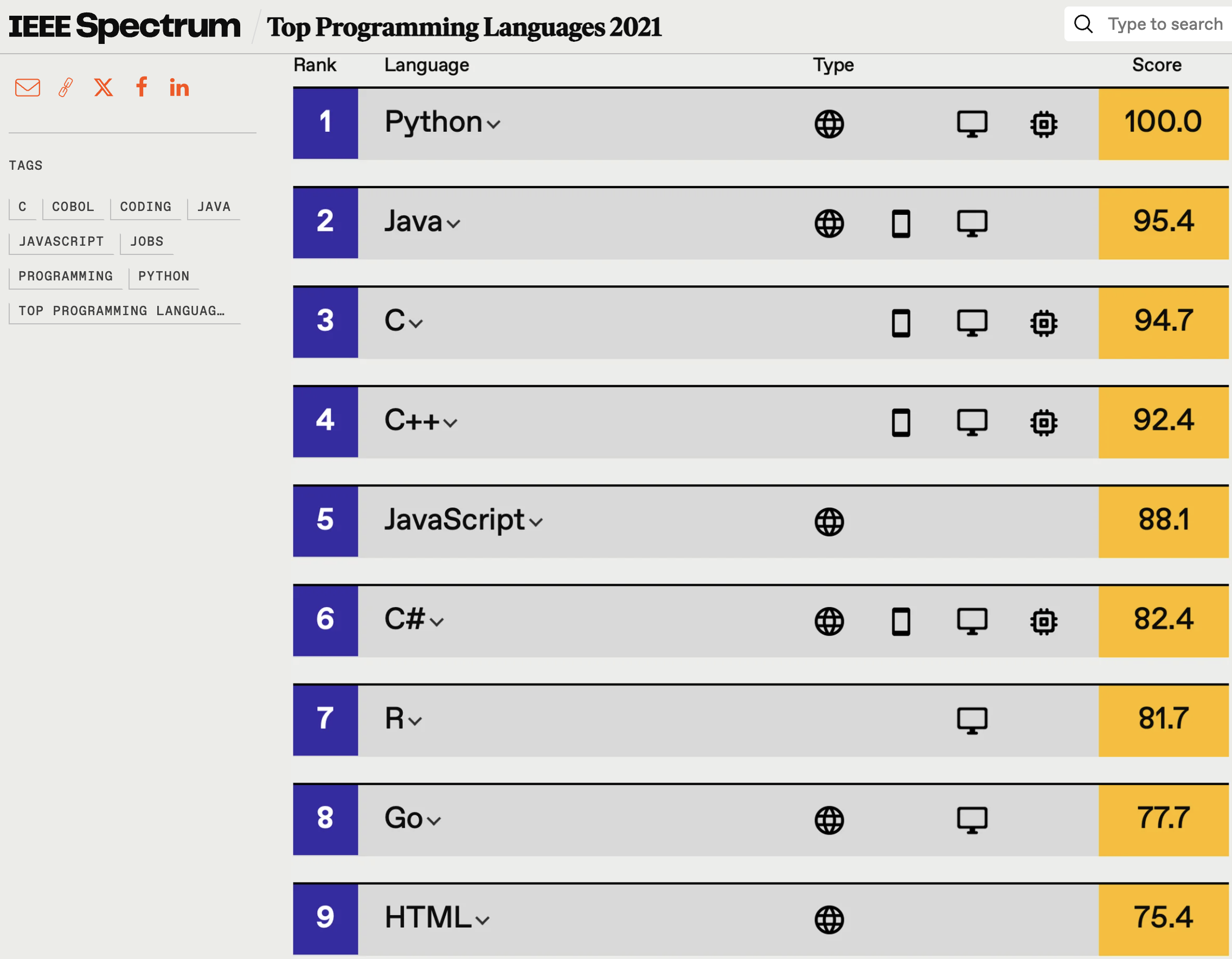
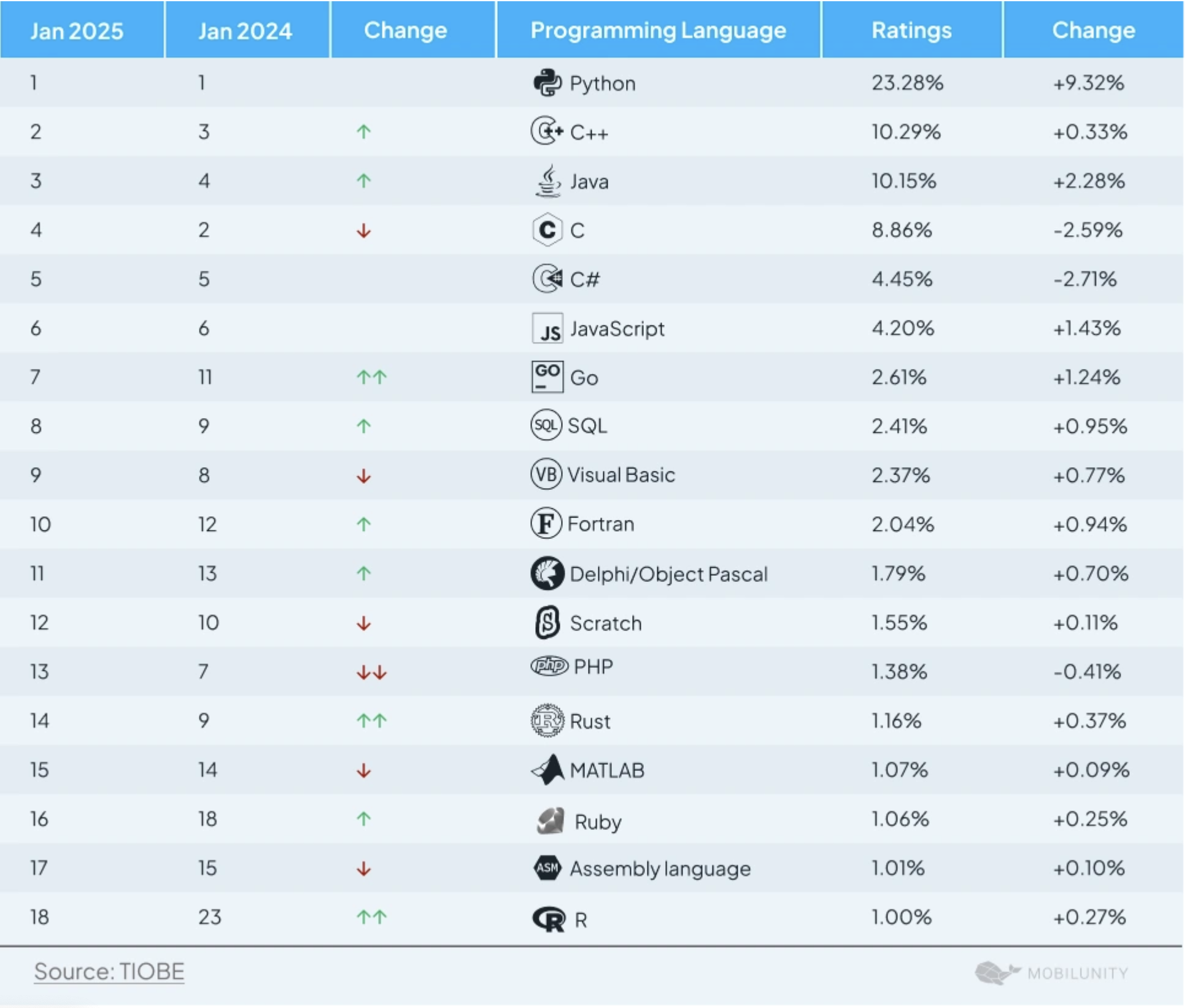
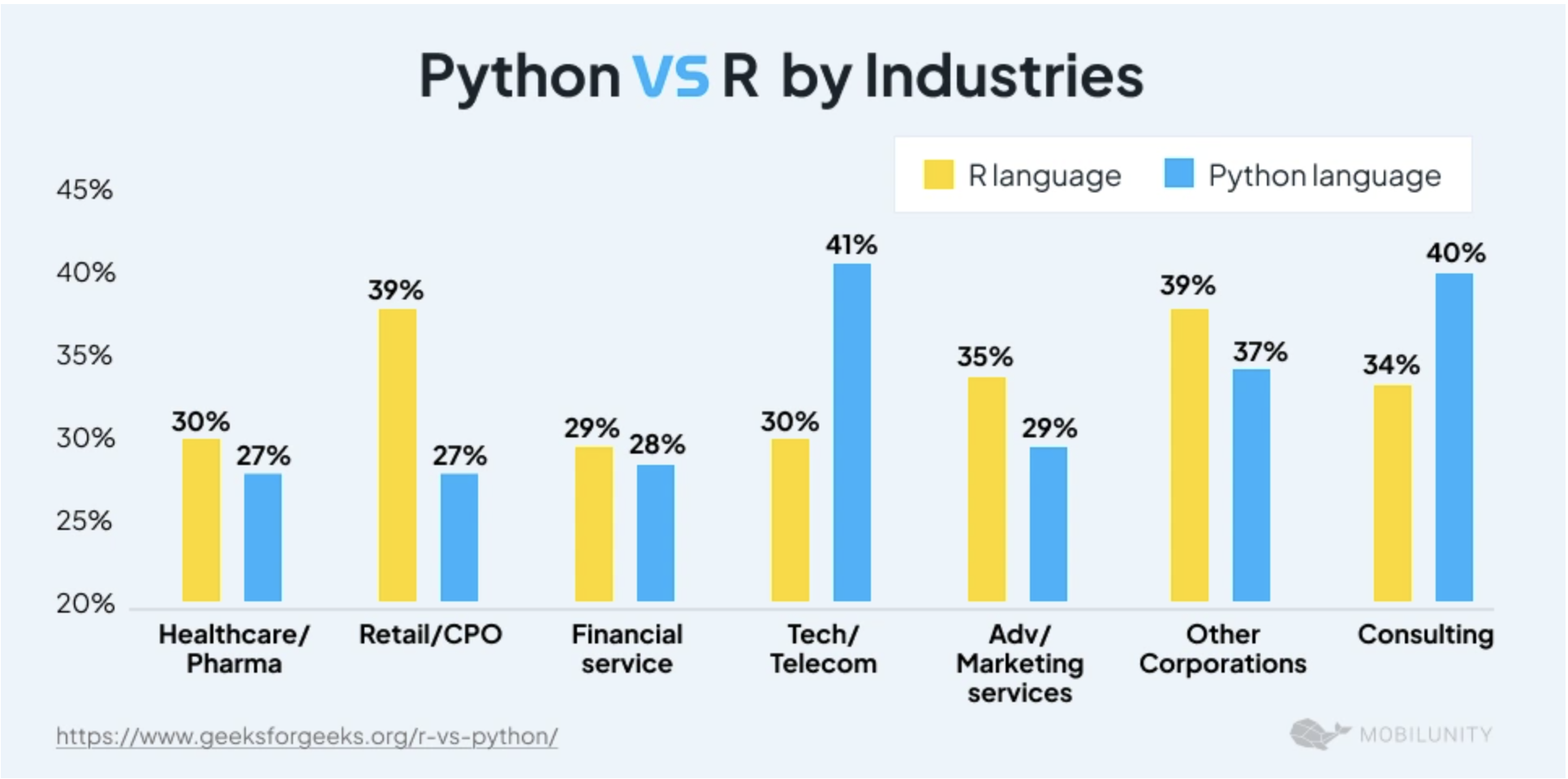
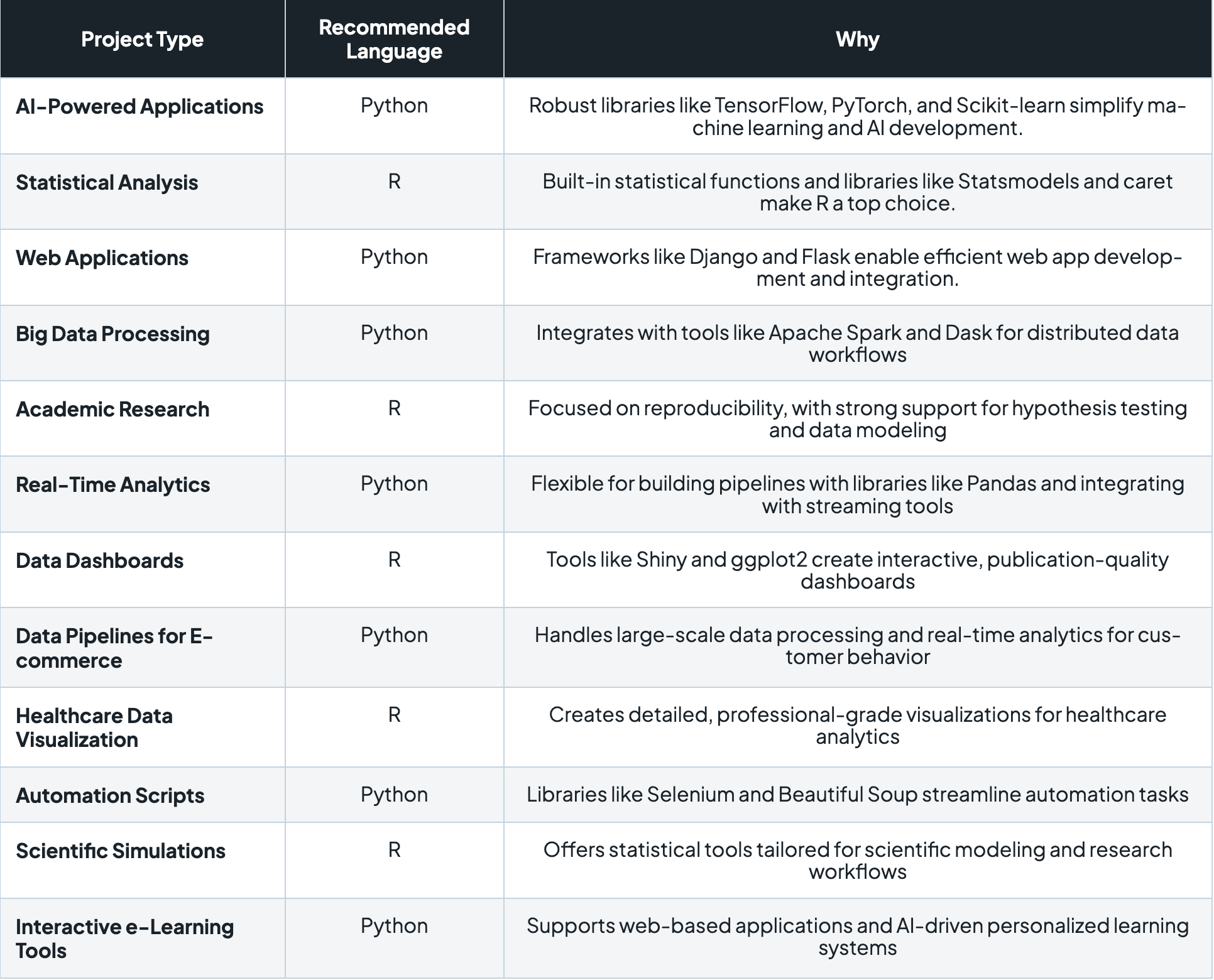
R vs Python
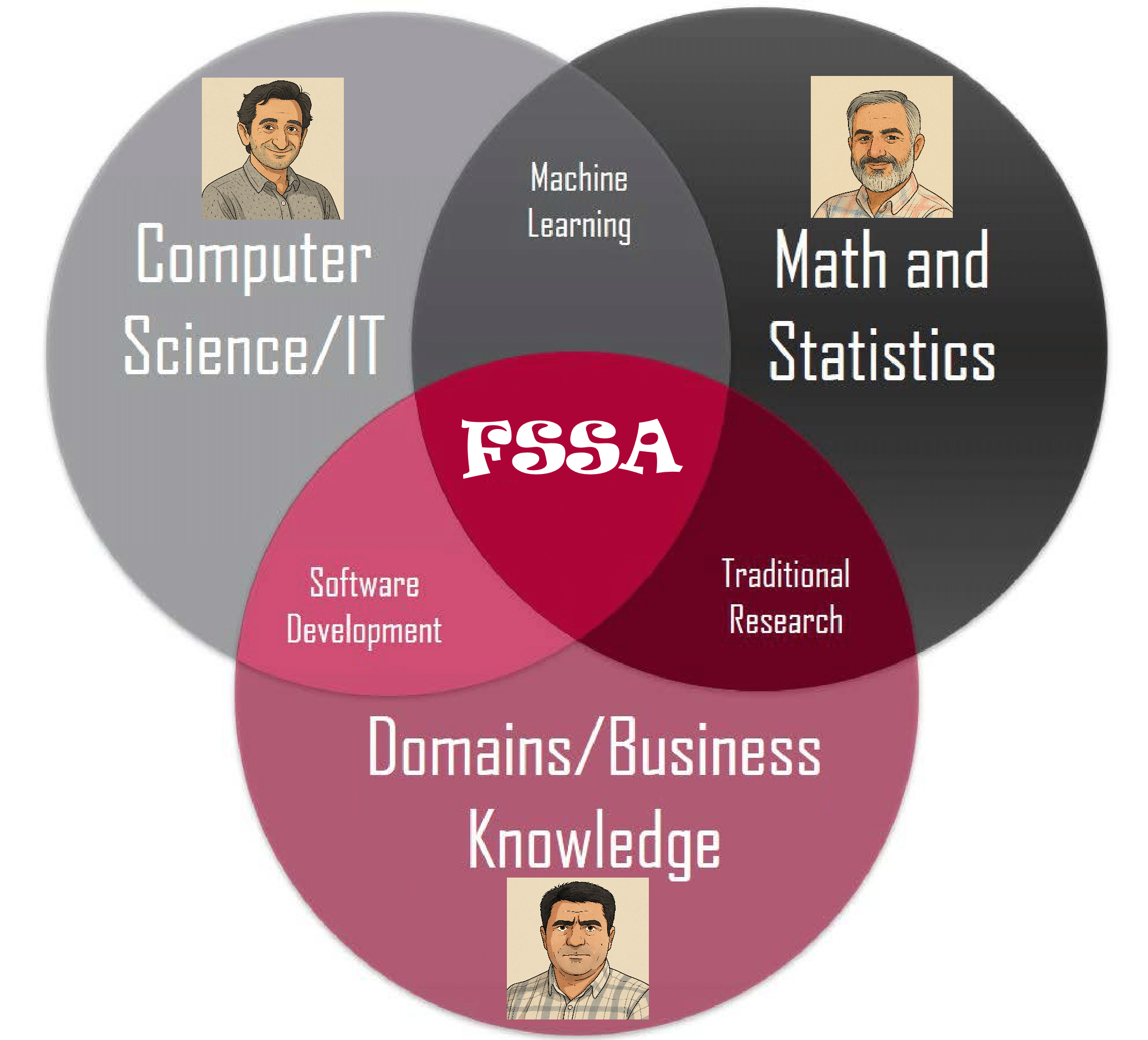
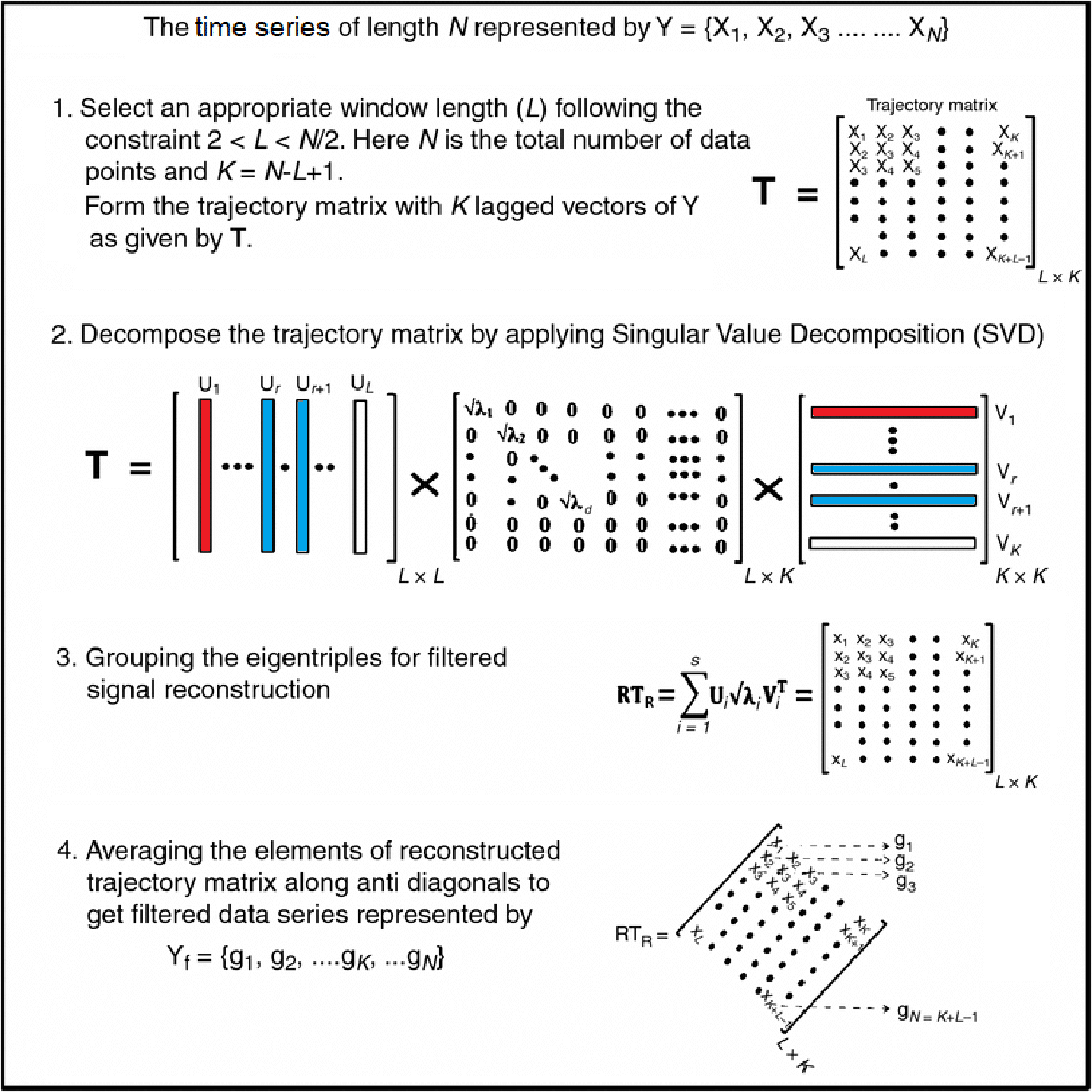
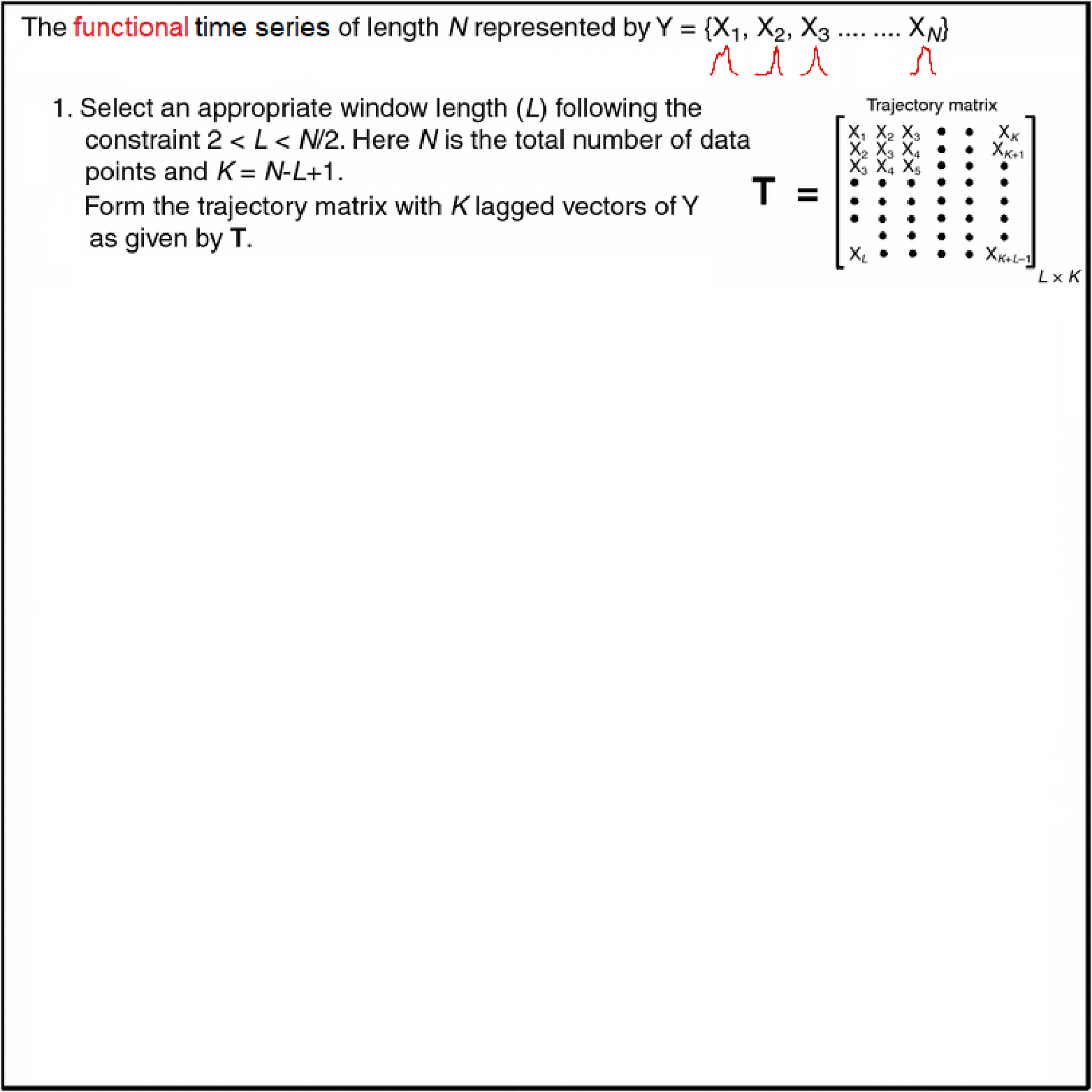
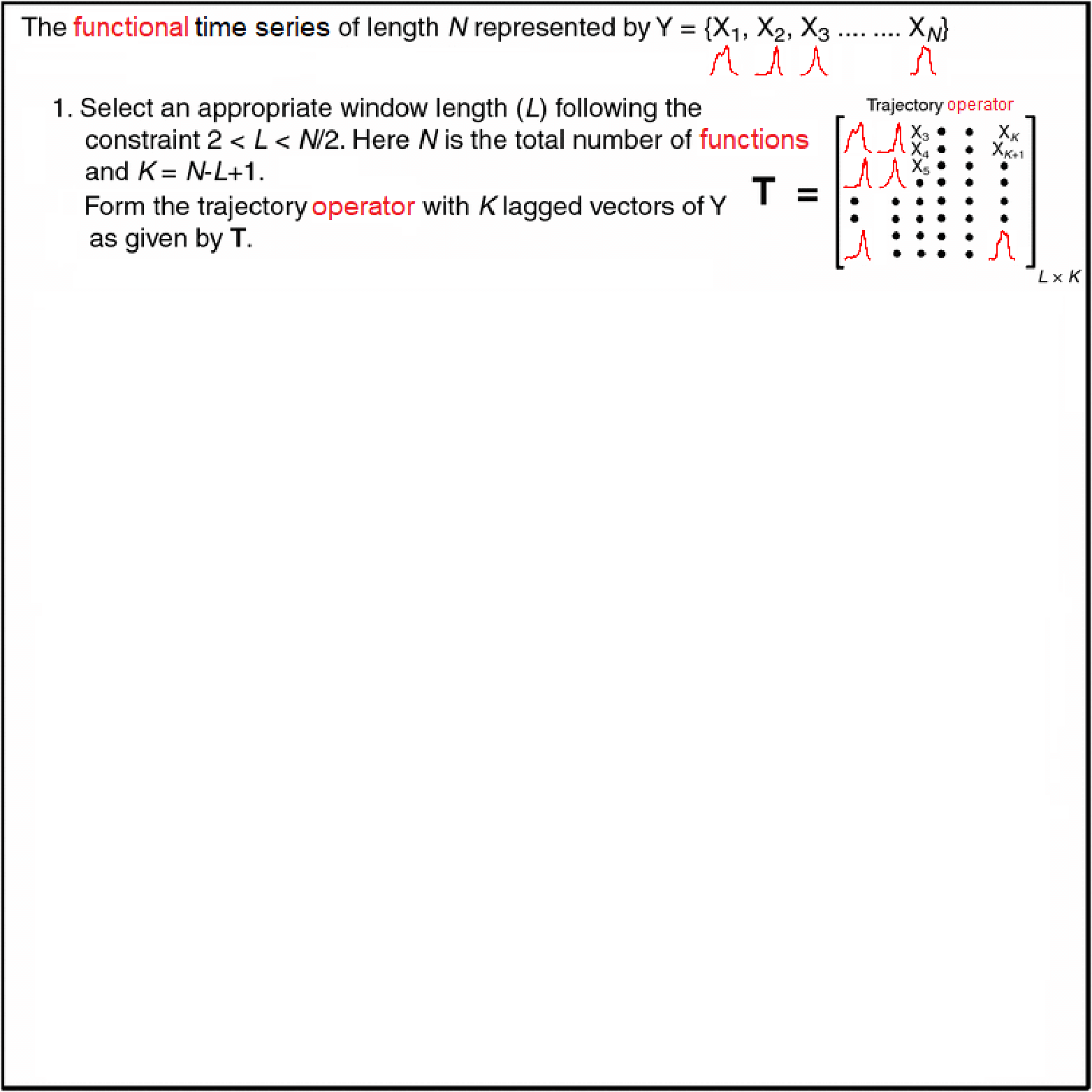
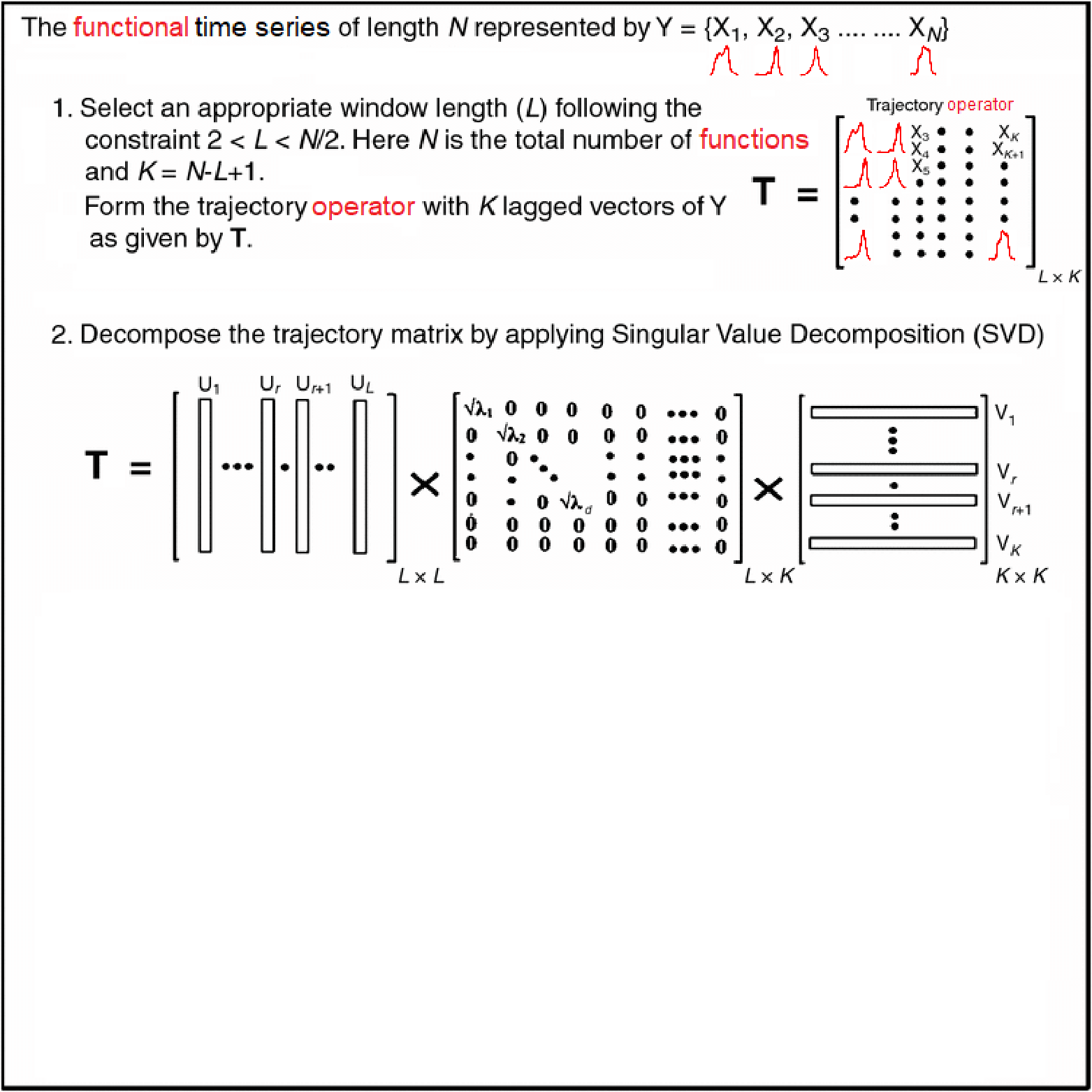
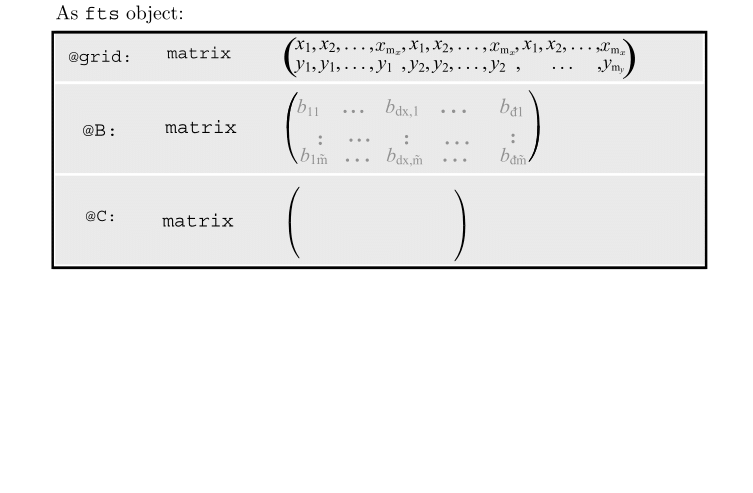
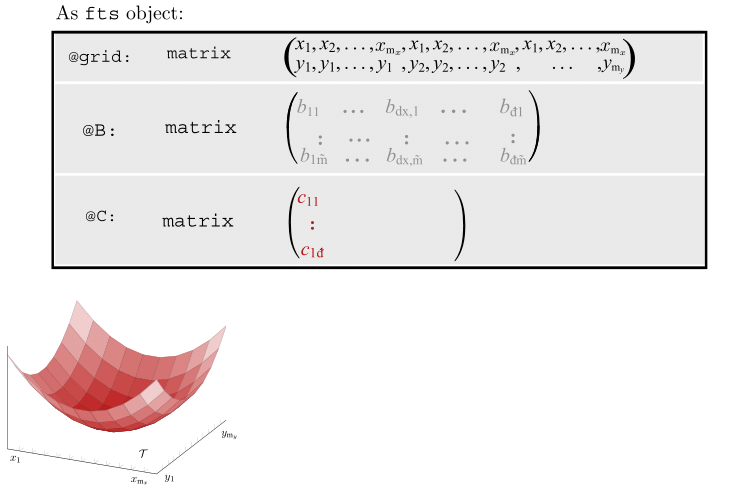
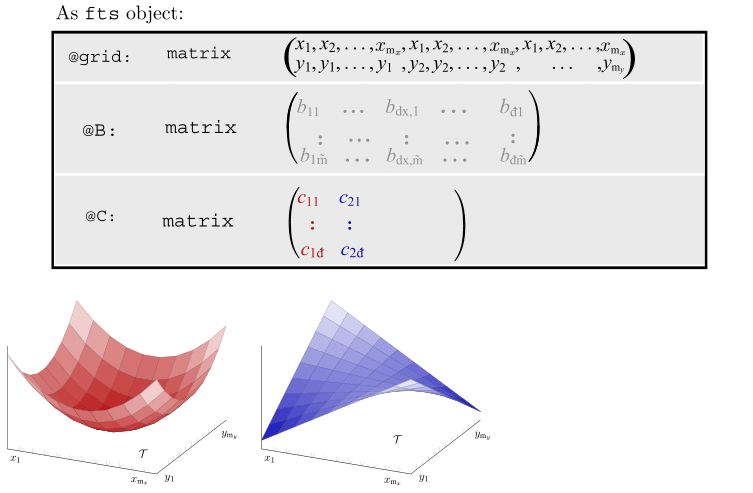
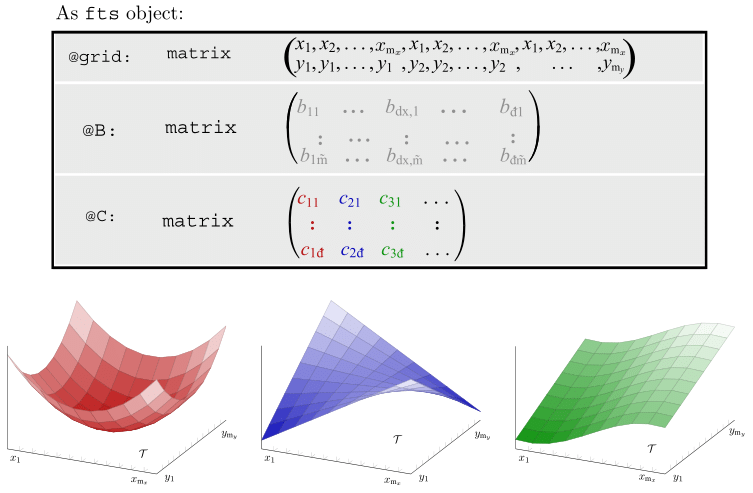
Motivating Example - Functional Singular Spectrum Analysis

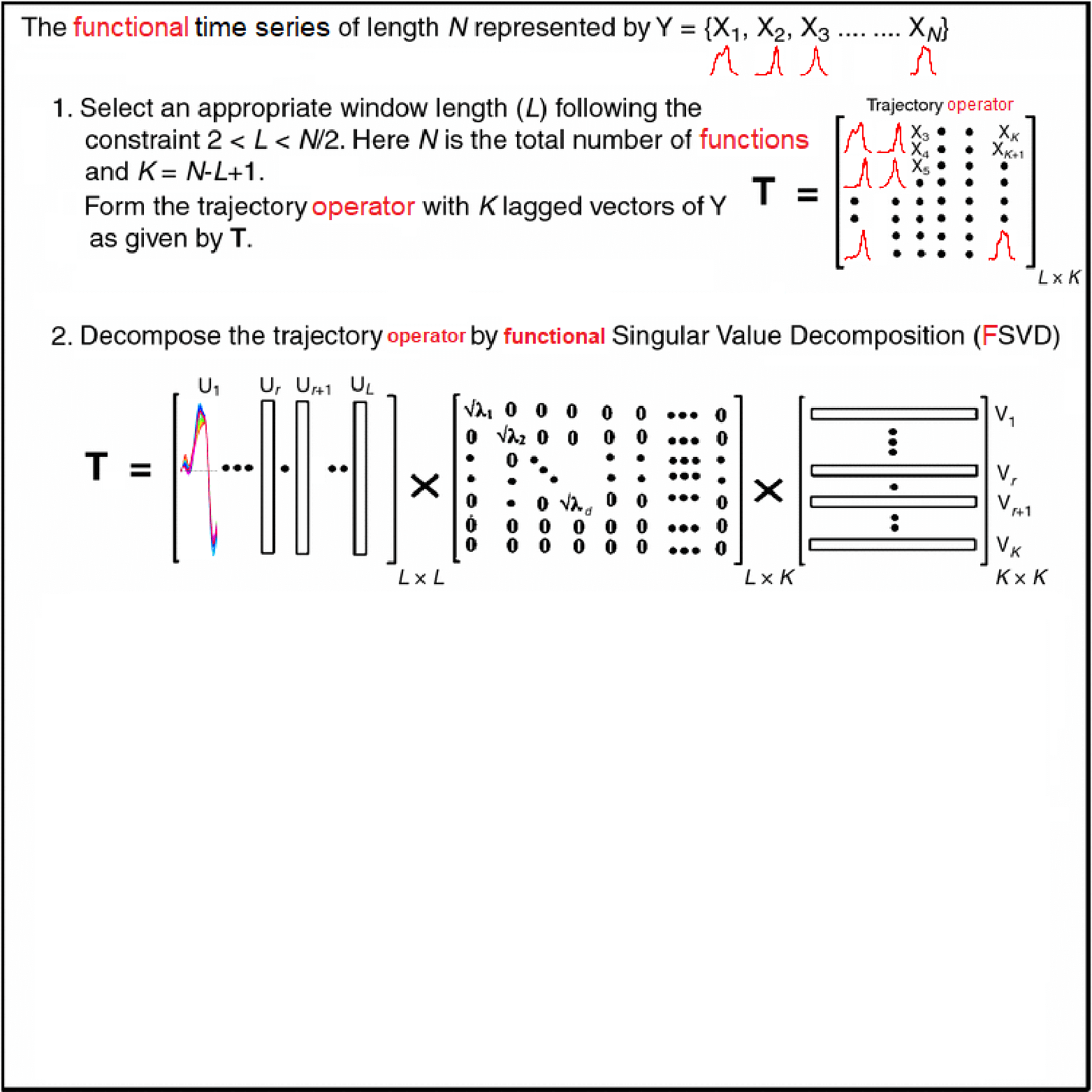
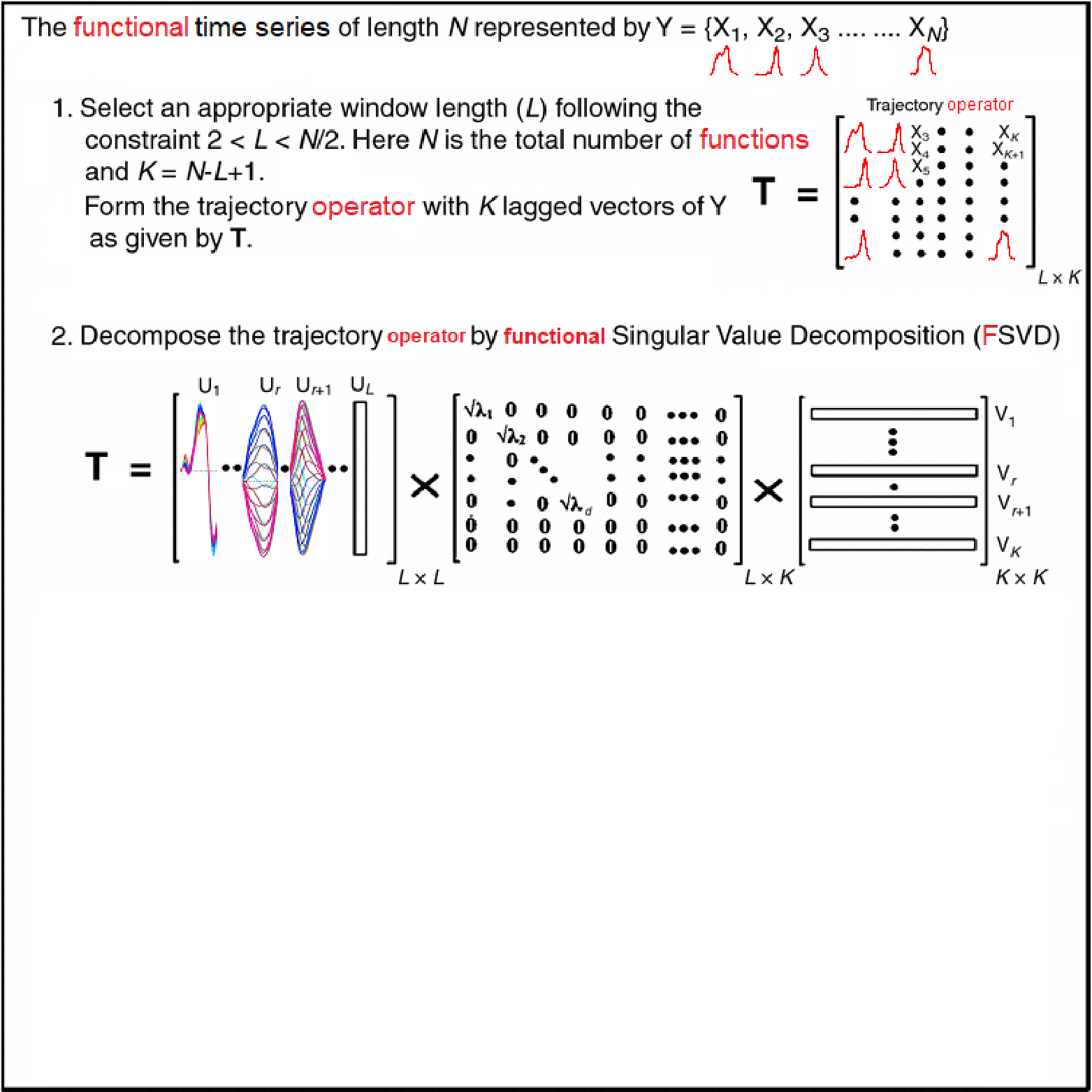
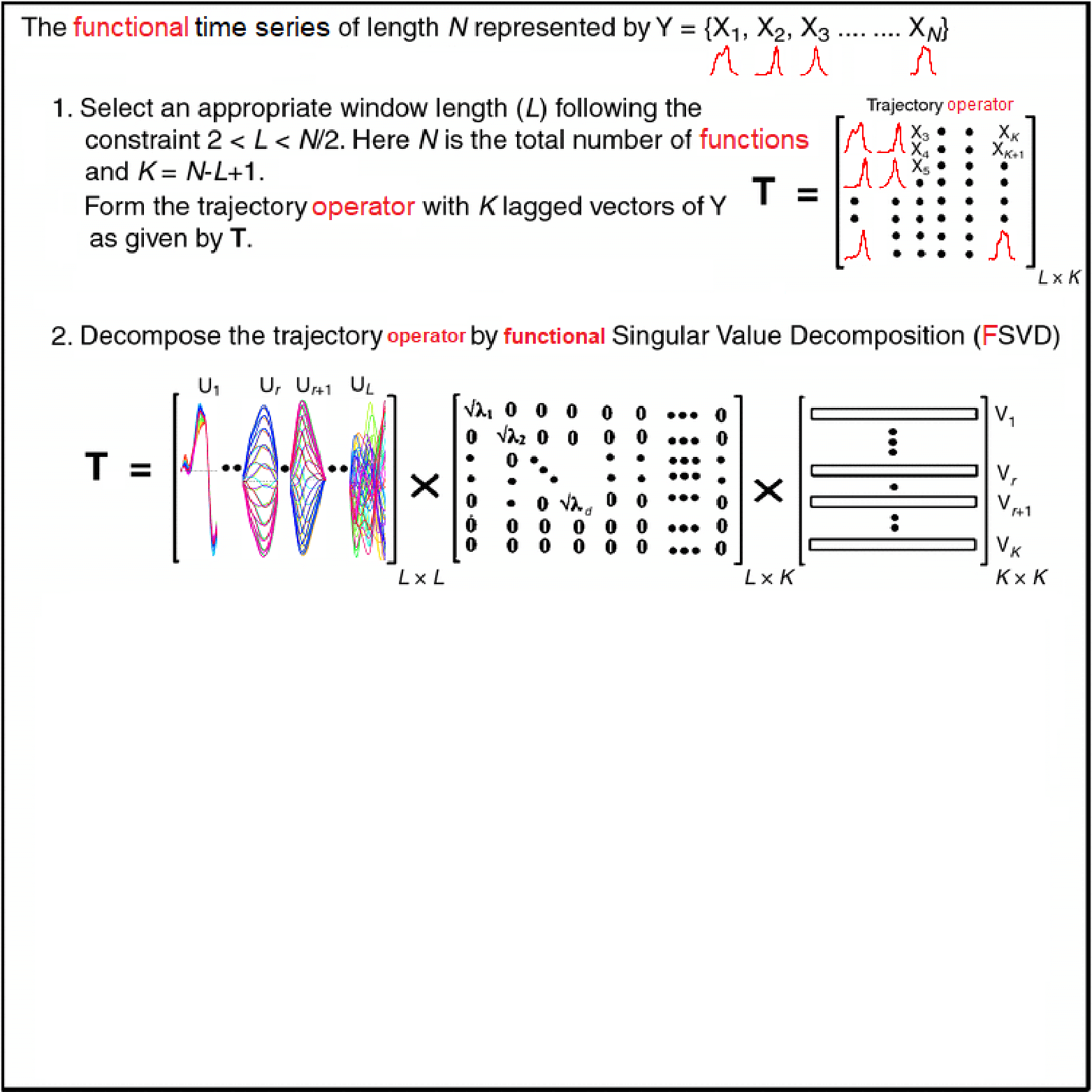
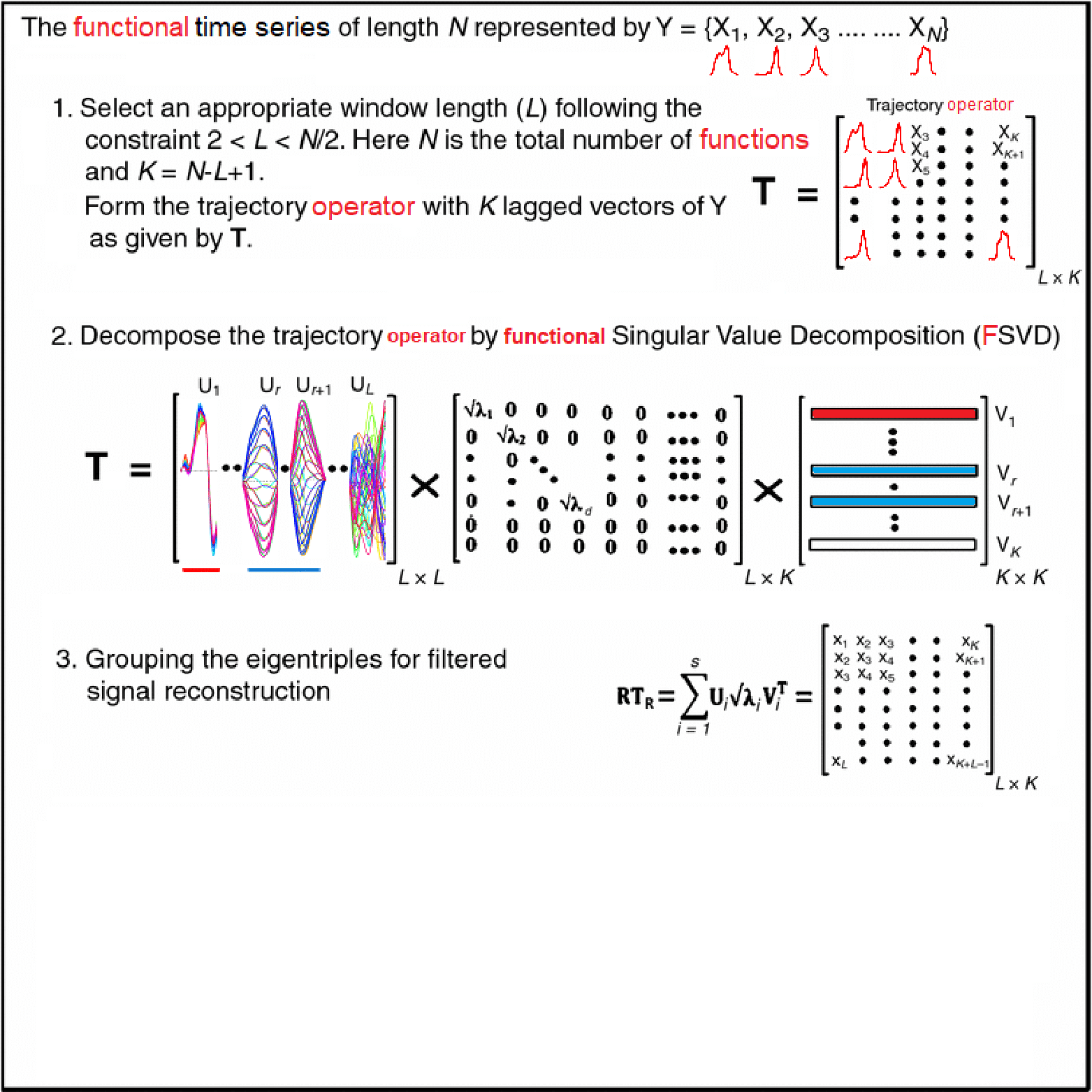
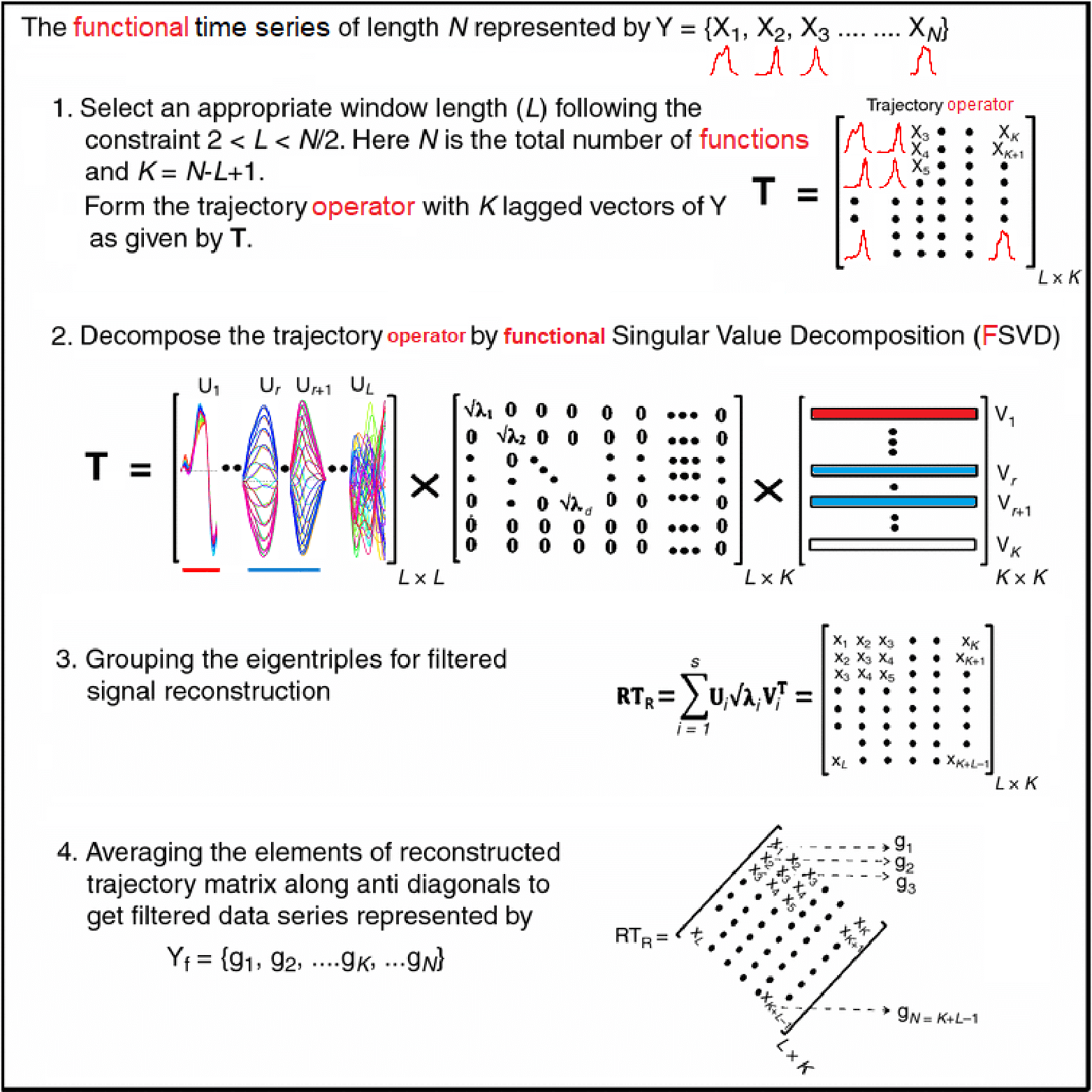
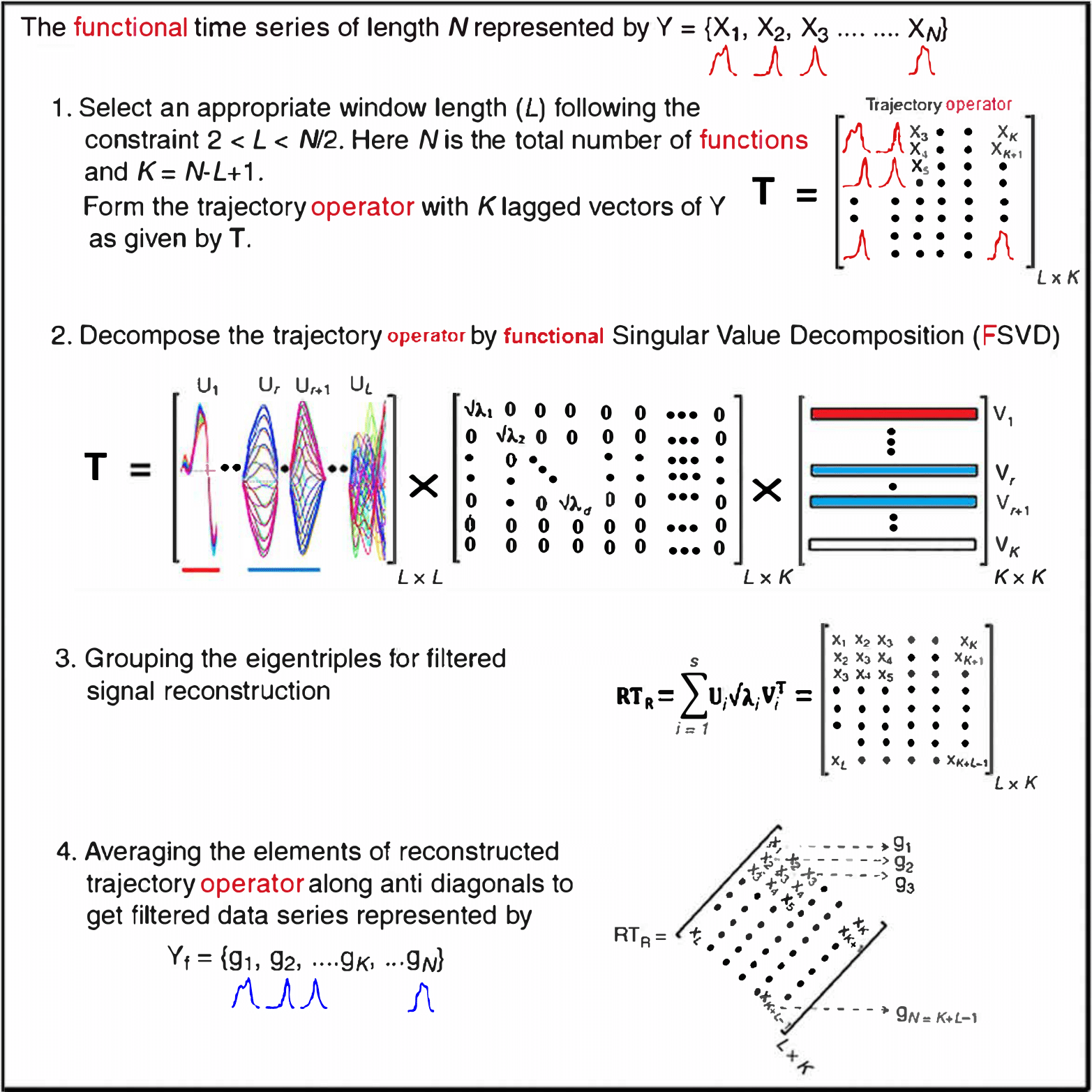
Outline
- Session 1. OOP in R
- Session 2. Web Apps with Shiny
- Session 3. Rcpp – High Performance with C++ in R
- Session 4. Python in R
- Session 5. Build Your Own R Package
- Session 6. CRAN Submission
- Session 7. GitHub Essentials
🧠 Session 1: OOP in R
Motivating Example - OOP
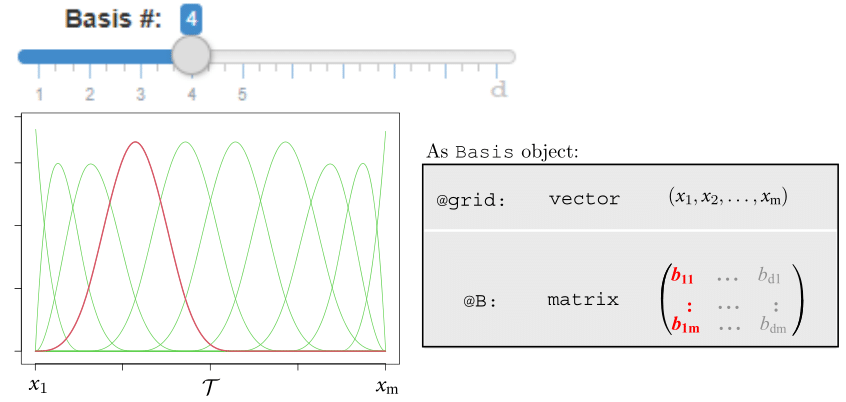
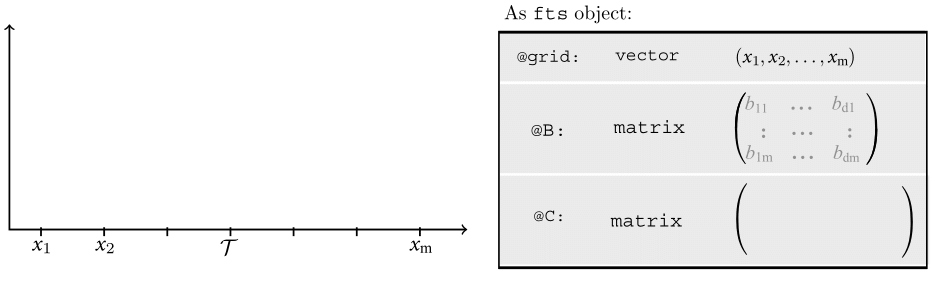
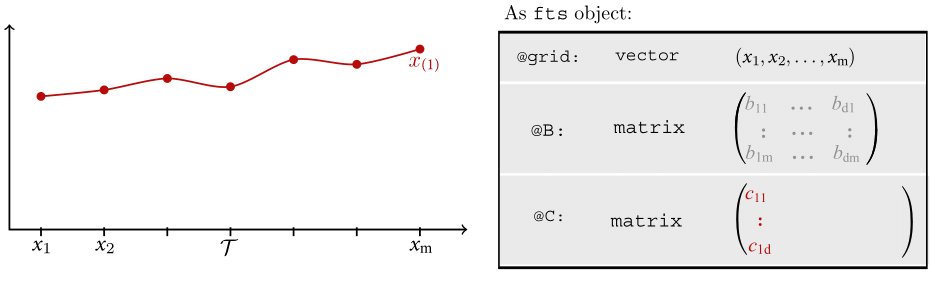
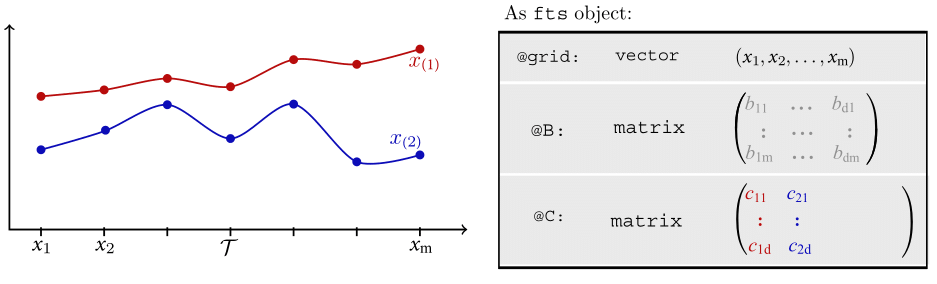
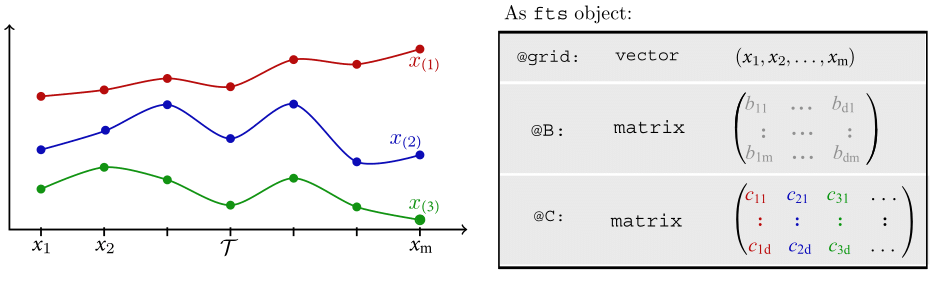
Motivating Example - OOP (Cont.)
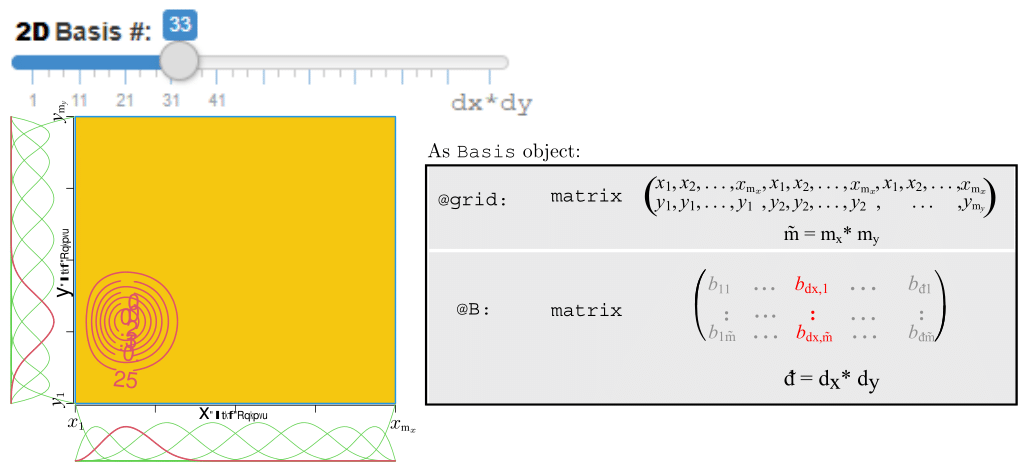
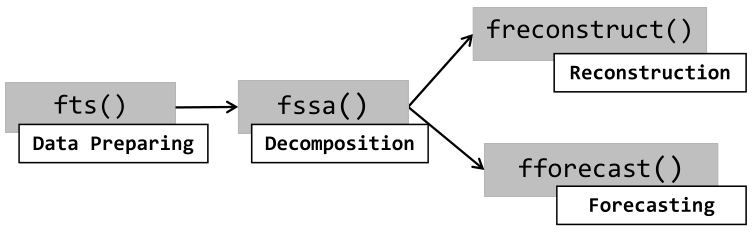

OOP in R
- 🧩 OOP Paradigms in R:
- Functional OOP:
plot(lm(...)) - Encapsulated OOP:
learner$train() - Modern tools:
mlr3,R6,S7
- Functional OOP:
- 🏗 OOP Systems:
- S3: Lightweight & informal — uses
UseMethod() - S4: Formal and type-safe — uses
setClass()and@ - R6: Mutable, used in apps like
shiny - S7: Newest and unified — multiple dispatch
- S3: Lightweight & informal — uses
🛠️ OOP in Practice
🔁 Comparison of Systems
| Sys. | Style | Used In | ✅ Pros | ⚠️ Cons |
|---|---|---|---|---|
| S3 | Functional | Base R | Simple, extensible | No schema, fragile |
| S4 | Formal | Bioconductor | Type-safe, robust | Verbose, complex |
| R6 | Encapsulated | Shiny, Keras | Fast, mutable | Weak type safety |
| S7 | Unified OOP | Modern dev | Combines S3 & S4 benefits | Still evolving |
Session 2:🌐 Web Apps with Shiny
FSSA - Shiny
Web Apps with Shiny
- ✨ Shiny = Interactive web apps in pure R (no HTML/JS!)
- 📦 Components:
- ui: Layout, inputs/outputs
- server: Logic, reactivity
- 🔌 Inputs & Outputs:
- Inputs: sliderInput(), textInput()
- Outputs: renderPlot(), renderText(), renderTable()
- 🧪 Use Cases:
- Dashboards, simulations, teaching tools
Shiny Concepts & Deployment in Workshop
- 🚀 Deployment Options:
shinyapps.io, Posit Connect, university servers- shinylive: deploy as static site (experimental)
- ⚡Reactivity System:
reactive(): memoized reactive expressionsisolate(): break reactive chainsobserveEvent(): trigger actions without outputs
- 🧠 Hands-on:
⚡ Session 3: Rcpp –
High Performance with C++ in R
Context & Motivation
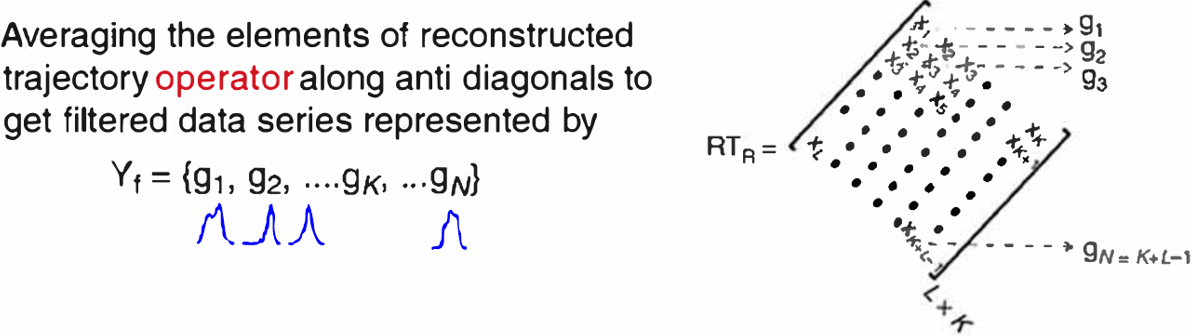
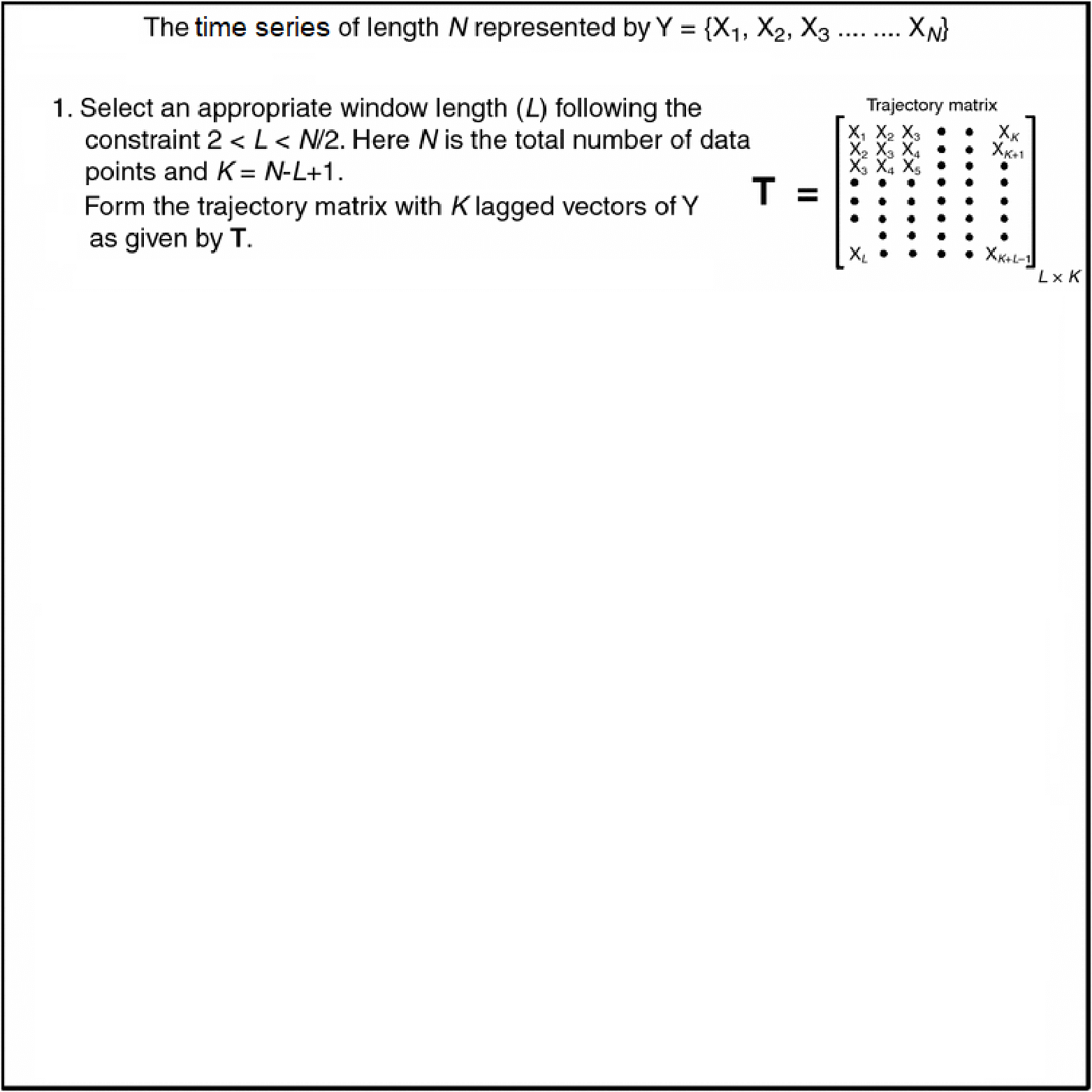
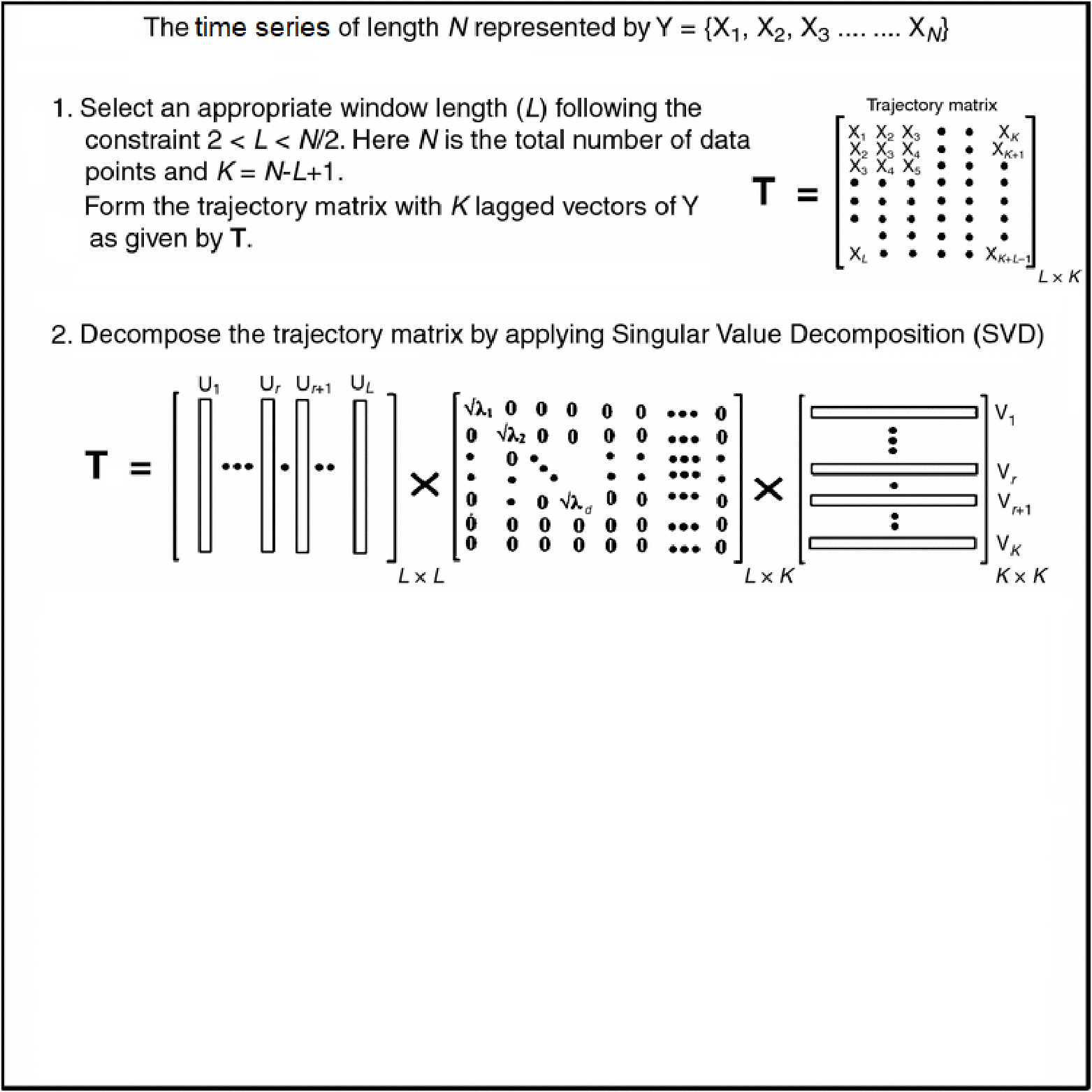
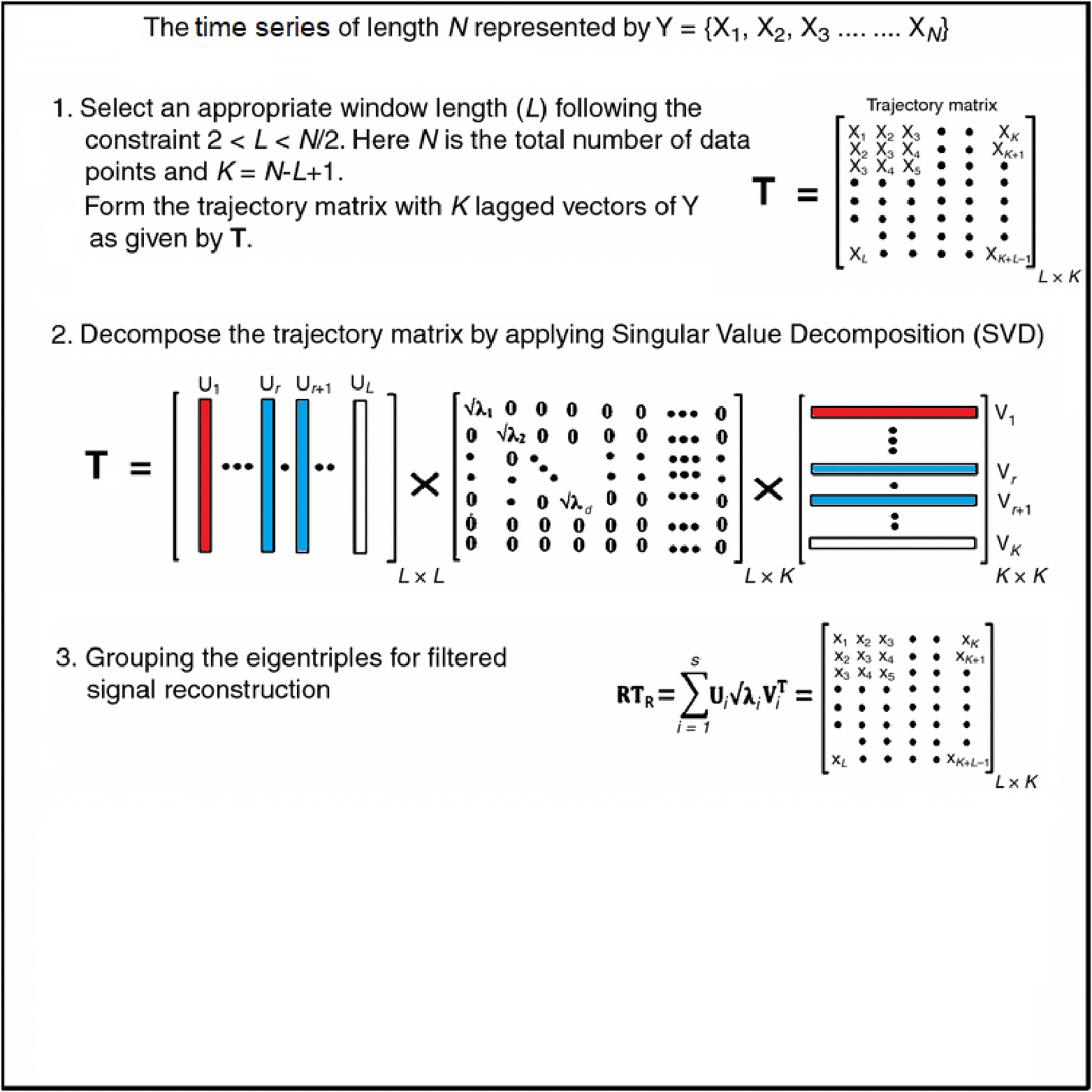
- Diagonal averaging is key in many algorithms (e.g. SSA reconstruction).
![]()
- Given any matrix \(G \in \mathbb{R}^{L\times K}\), we want \[ \scriptsize{y_t = \frac{1}{\#\{(i,j):\,i+j-1 = t\}} \sum_{i+j-1 = t} G_{i,j}, \quad t = 1,\dots, L+K-1.} \]
- Pure‑R implementation uses two nested loops over \(i\) and \(j\).
Pure R Code
Rcpp Code
library(Rcpp)
cppFunction('
NumericVector hankelize_rcpp(NumericMatrix X) {
int L = X.nrow();
int K = X.ncol();
int N = L + K - 1;
NumericVector result(N);
NumericVector count(N);
for(int i = 0; i < L; i++){
for(int j = 0; j < K; j++){
result[i + j] += X(i,j);
count[i + j] += 1;
}
}
for(int i = 0; i < N; i++){
result[i] /= count[i];
}
return result;
}')Benchmark
library(microbenchmark)
# Large random matrix (e.g., 100 x 1000)
set.seed(123)
X_large <- matrix(runif(1e5), nrow = 100, ncol = 1000)
bench <- microbenchmark(
R_version = hankelize_R(X_large),
Rcpp_version = hankelize_rcpp(X_large),
times = 10
)
print(bench)
># Unit: microseconds
># expr min lq mean median uq max neval
># R_version 21225.1 21550.8 26604.53 25180.35 30969.3 34455.1 10
># Rcpp_version 176.9 177.8 299.87 194.50 253.7 1161.3 10- Dramatic speedup: ~89× faster.
- Pattern can be applied to many \(O(n^2)\) tasks in time series, spatial stats, and beyond.
🔬 Rcpp in Action
- 🚀 Rcpp bridges R and C++ for high-performance computing.
- 🏁 Use cases:
- Speed up loops, recursion, and matrix operations
- Access C++ libraries and STL
- Integrate C++ in R packages
🐍 4. Python in R
🔁 Example: Use Python from R
🐍 Native Python
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
from sklearn.neural_network import MLPClassifier
iris = load_iris()
X, y = iris.data, iris.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
mlp = MLPClassifier(hidden_layer_sizes=(10,))
mlp.fit(X_train, y_train)
y_pred = mlp.predict(X_test)📦 R with reticulate
library(reticulate)
sklearn <- import("sklearn")
model_selection <- sklearn$model_selection
neural_network <- sklearn$neural_network
X <- as.matrix(iris[, 1:4])
y <- as.integer(iris$Species) - 1
train_test <- model_selection$train_test_split(X, y, test_size = 0.2, random_state = 42)
X_train <- train_test[[1]]
X_test <- train_test[[2]]
y_train <- train_test[[3]]
y_test <- train_test[[4]]
mlp <- neural_network$MLPClassifier(hidden_layer_sizes = tuple(10L))
mlp$fit(X_train, y_train)
y_pred <- mlp$predict(X_test)💡 Why Call Python in R?
- 🔄 Avoid rewriting code from one language to another
- 🔁 Call shared models or preprocessing functions written in Python
- 📦 Reuse Python packages or APIs within your R workflow
- 📤 Publish reproducible reports that include both R and Python code
What is reticulate?
- R package for interoperability with Python
- Maintains a shared Python session accessible from R
- Automatic conversion of many R types to Python and vice versa
- Supports inline Python code in R Markdown and R scripts
🎥 Demo: Python from R in RStudio
📦 5. Build Your Own R Package
💡 Motivations to Package Your Code
🧰 Why build a package?
- ♻️ Reuse and organize reusable code chunks
- 📖 Provide documentation and examples
- 🧪 Add unit tests to ensure reliability
- 🌐 Distribute:
- via CRAN for wide access
- via GitHub for collaborative development
- or keep it private for internal use
🏗 Package Structure Overview
mypkg/
├── DESCRIPTION # 📋 Package metadata
├── NAMESPACE # 🔁 Exported functions & imports
├── R/ # 📂 Your R functions
└── man/ # 📚 Auto-generated documentation💡 Other folders you may add:
- 📂 tests/ → Unit tests (testthat)
- 📂 vignettes/ → Long-form docs
- 📂 data/ → Internal or example datasets
🎥 Demo: Rfssa Package
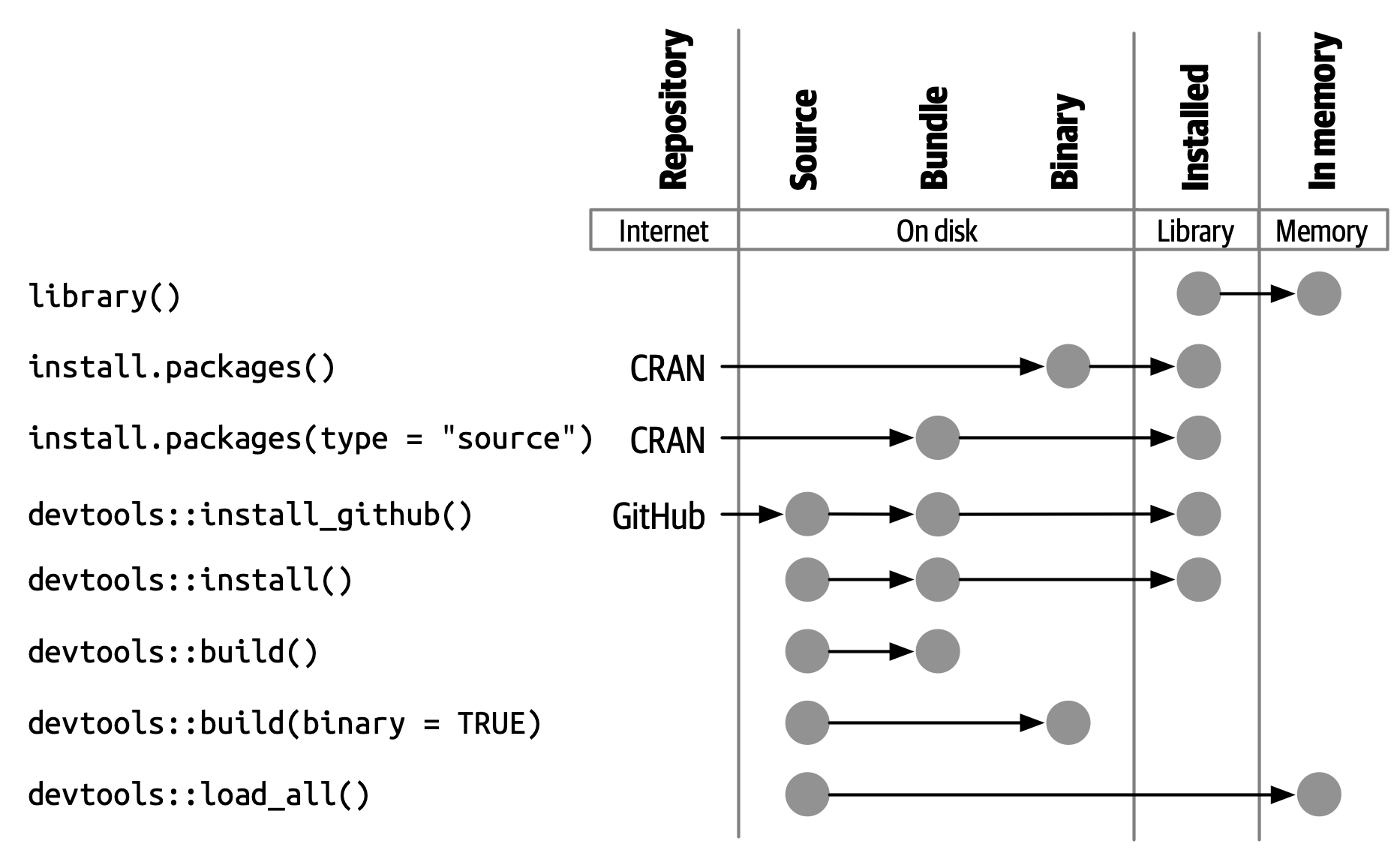
🛠 Package States in R
R packages transition through five development states:
- 🗂️ Source: your raw package folder
- 📦 Bundled: compressed .tar.gz for sharing
- 🧱 Binary: platform-specific precompiled version
- 📚 Installed: available in your R library
- 🧠 In-Memory: actively loaded via library()
🔁 Transitioning Between States
6. 📤 CRAN Submitting
🚀 Submitting to CRAN
- 📦 CRAN = Comprehensive R Archive Network
- 🎯 Goal: Make your R package public, discoverable, and installable
- ✔ Must pass
R CMD checkwith:- ❌ No ERRORs
- ⚠️ No WARNINGs
- 📝 Minimal and justifiable NOTEs
- 📤 Submit with:
devtools::release()
📩 Confirmation Email (After Submission)
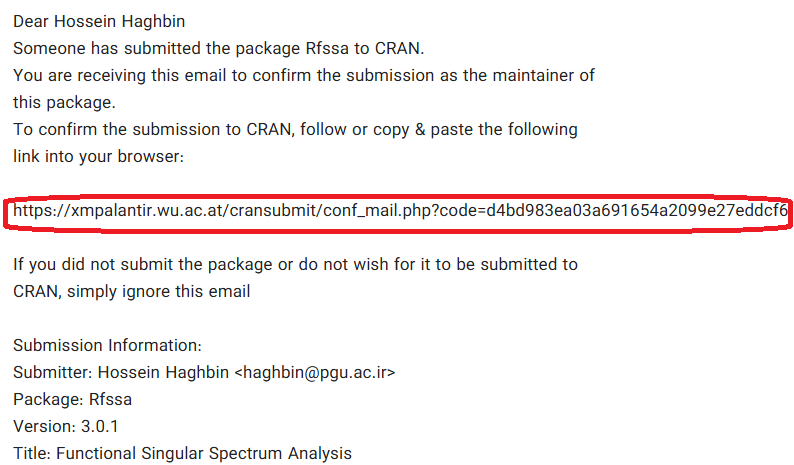
📬 Submission Confirmed Email

⏳ Pending Manual Inspection Email
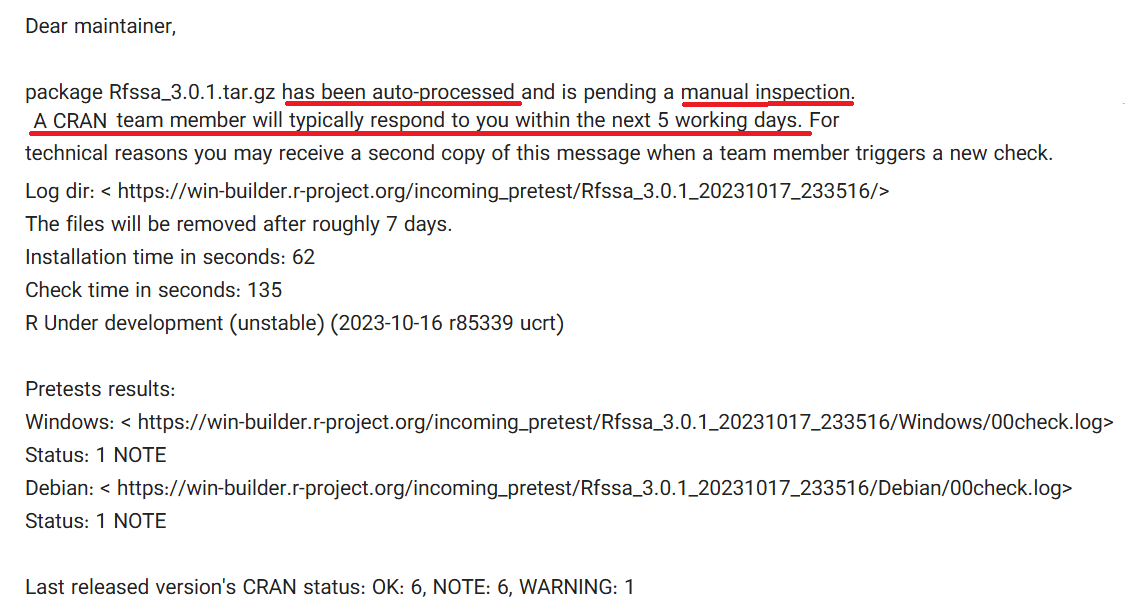
✅ Package Initially Accepted Email
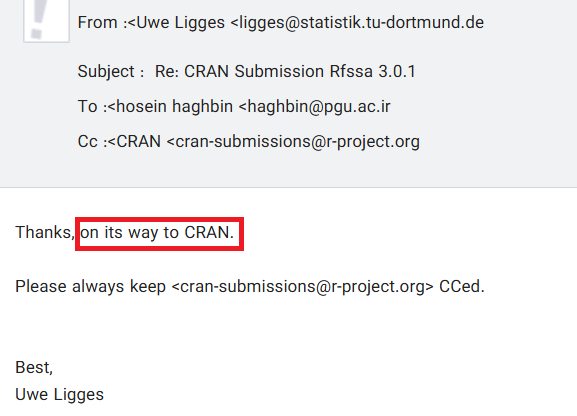
🎉 Good news!
📦 Package Built on CRAN Email
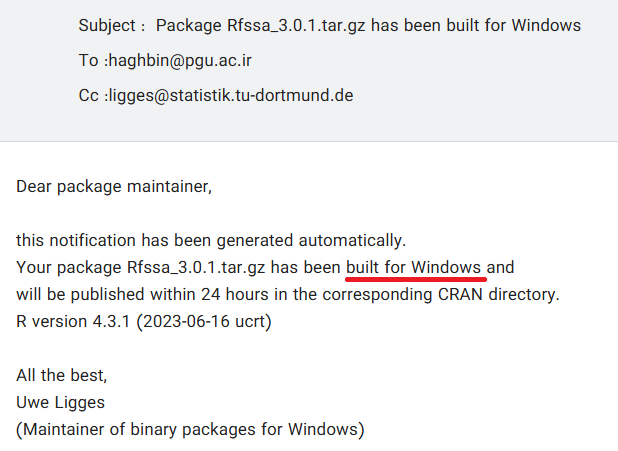
🧱 CRAN has built binaries for major platforms (Windows, …).
🌐 CRAN Page Created
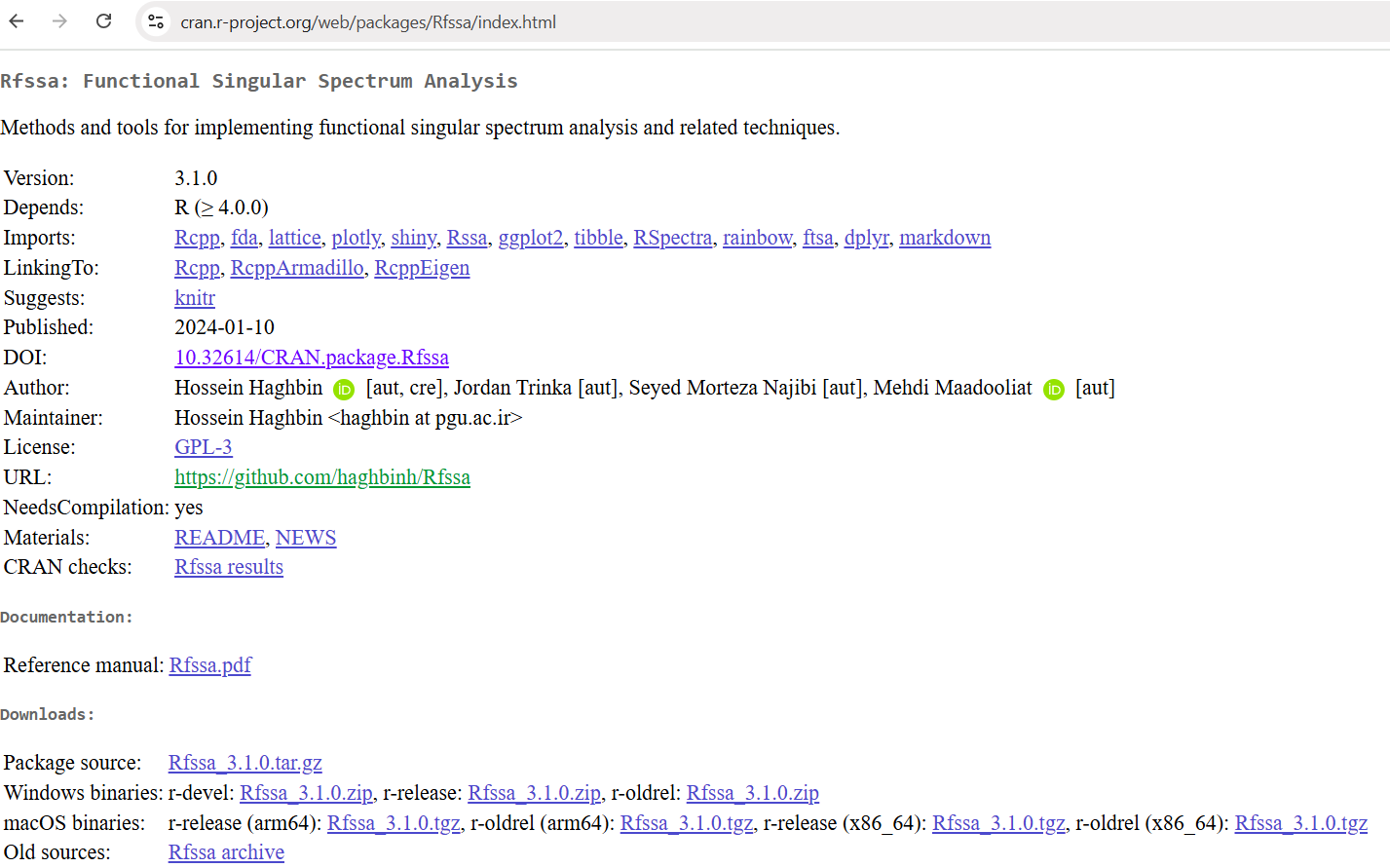
🌍 Your package is now live! Searchable and installable via:
7. 📤 GitHub Essentials
📬 Example: Rfssa Package Github
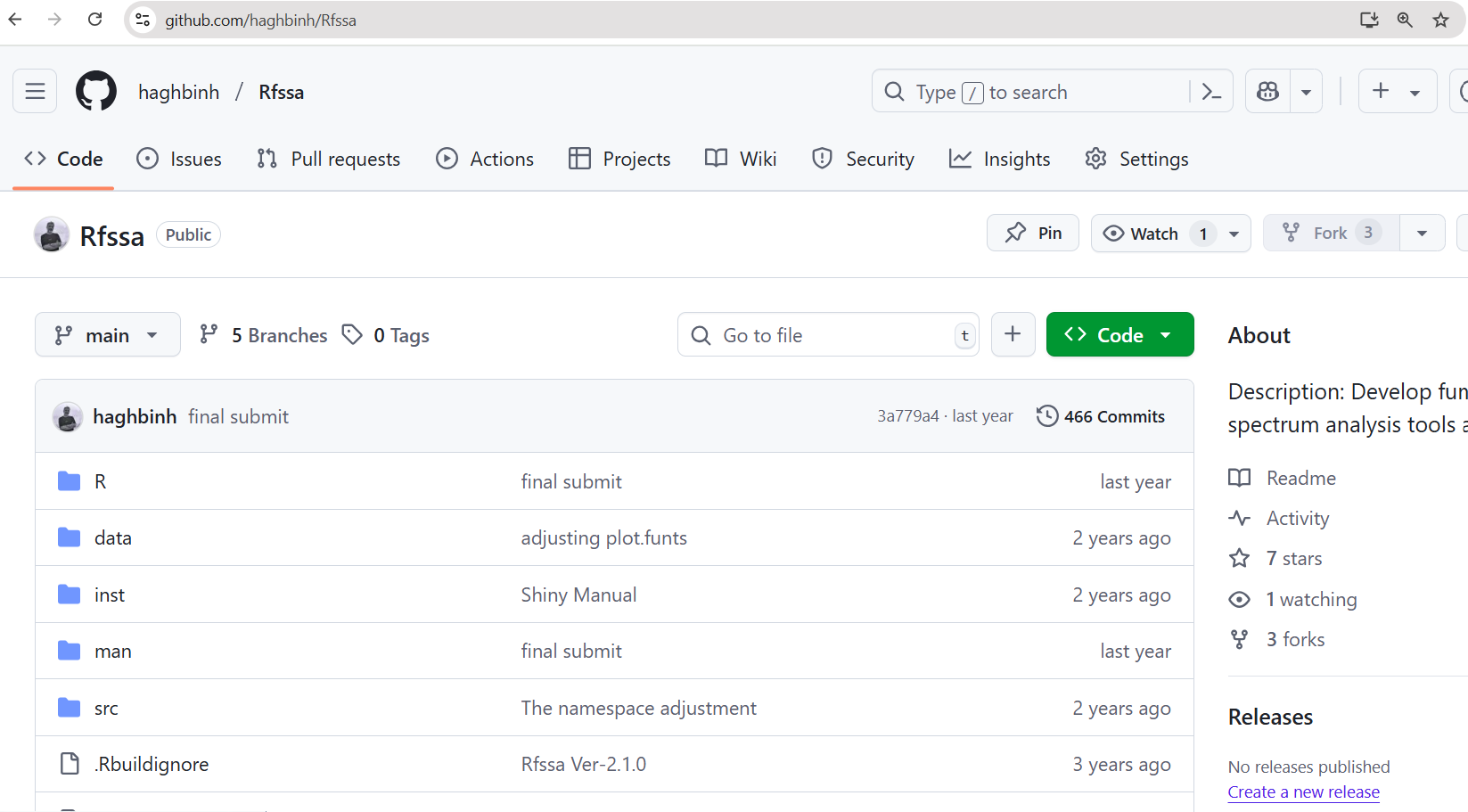
🌍 Why Use Git & GitHub?
- 🔁 Version Control: Track and manage changes in code
- 🤝 Collaboration: Work with others without overwriting files
- 🧪 Reproducibility: Restore older versions when bugs occur
- 🔄 Open Source: Share your package with the world
🧰 Git = Local tracking tool
☁️ GitHub = Online hosting and teamwork platform
🔁 Daily Git Workflow in RStudio
🎯 A typical work cycle for package developers:
| Step | Description |
|---|---|
| ✏️ Edit | Make changes to .R or .Rmd files |
| ✅ Stage | Select files to track in Git tab |
| 💬 Commit | Save with a message (e.g., “Fix bug”) |
| 🚀 Push | Send to GitHub |
🎥 Demo: Rfssa Github Package
🙏 Thank you!
Questions & Discussion